初心者ながらKaggleに挑戦した時のコードを備忘録として残しておきます。
・Kaggle関係の記事
Kaggleのタイタニックに挑戦してみた(その1)
Kaggleのタイタニックに挑戦してみた(その2)
Kaggleで書いたコードの備忘録その1(ここ)
Kaggleで書いたコードの備忘録その2~自然言語処理まとめ~
KaggleタイタニックでNameだけで予測精度80%超えた話(BERT)
1.データ
import
全体的に使うライブラリです。
各項目で使うライブラリはそちら側でimportを記載しています。
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
使用するデータ
Kaggleチュートリアルのタイタニックを使います。
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
df_test = pd.read_csv("/kaggle/input/titanic/test.csv")
学習データとテストデータをまとめて管理する
- マージ
# データをマージ
df_test["Survived"] = np.nan
df = pd.concat([df_train, df_test], ignore_index=True, sort=False)
print(df_train.shape) # (891, 12)
print(df_test.shape) # (418, 12)
print(df.shape) # (1309, 12)
※目的変数(Survived)のテスト側の値はnanになります
- 分割
df_train = df[df["Survived"].notnull()]
df_test = df[df["Survived"].isnull()]
print(df_train.shape) # (891, 12)
print(df_test.shape) # (418, 12)
数値データの要約統計量を確認したい
print(df[["Age", "Fare"]].describe())
Age Fare
count 1046.000000 1308.000000
mean 29.881138 33.295479
std 14.413493 51.758668
min 0.170000 0.000000
25% 21.000000 7.895800
50% 28.000000 14.454200
75% 39.000000 31.275000
max 80.000000 512.329200
カテゴリデータの要約統計量を確認したい
print(df[["Sex"]].describe(include="O"))
Sex
count 1309
unique 2
top male
freq 843
※欠損値があるとエラーが出るので注意
欠損値を確認したい
- 方法1
print(df.isnull().sum())
PassengerId 0
Survived 418
Pclass 0
Name 0
Sex 0
Age 263
SibSp 0
Parch 0
Ticket 0
Fare 1
Cabin 1014
Embarked 2
is_train 0
dtype: int64
- 方法2
print(df.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1309 entries, 0 to 1308
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 1309 non-null int64
1 Survived 891 non-null float64
2 Pclass 1309 non-null int64
3 Name 1309 non-null object
4 Sex 1309 non-null object
5 Age 1046 non-null float64
6 SibSp 1309 non-null int64
7 Parch 1309 non-null int64
8 Ticket 1309 non-null object
9 Fare 1308 non-null float64
10 Cabin 295 non-null object
11 Embarked 1307 non-null object
12 is_train 1309 non-null int64
dtypes: float64(3), int64(5), object(5)
memory usage: 133.1+ KB
方法2は欠損値以外の情報も表示されます。
2.データの可視化
カテゴリデータを表示したい(1変数)
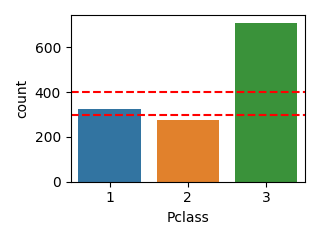
def plot_category(df, column, scale=1.0, line=None):
plt.figure(figsize=(6.4*scale, 4.8*scale))
# 表示メイン
sns.countplot(df[column])
if line is not None:
if type(line) == list:
for l in line:
plt.axhline(y=l, color="red", linestyle="--")
else:
plt.axhline(y=line, color="red", linestyle="--")
plt.tight_layout()
plt.show()
# 表示例
plot_category(df, "Pclass", scale=0.5, line=[300, 400])
数値データを表示したい(1変数)
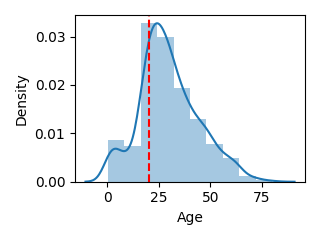
def plot_float(df, column, bins=10, scale=1.0, line=None):
plt.figure(figsize=(6.4*scale, 4.8*scale))
# 表示メイン
sns.distplot(df[column], kde=True, rug=False, bins=bins)
if line is not None:
if type(line) == list:
for l in line:
plt.axvline(x=l, color="red", linestyle="--")
else:
plt.axvline(x=line, color="red", linestyle="--")
plt.tight_layout()
plt.show()
# 表示例
plot_float(df, "Age", scale=0.5, line=20)
※binsはヒストグラムの分割数です
カテゴリデータに対してカテゴリデータを比較したい(2変数)
plot表示
- シンプル
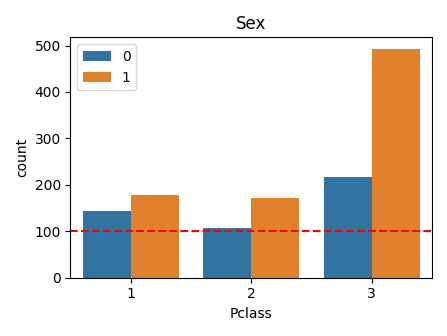
def plot_category_category(df, column1, column2, scale=1.0, line=None):
plt.figure(figsize=(6.4*scale, 4.8*scale))
# 表示メイン
sns.countplot(column1, hue=column2, data=df)
if line is not None:
if type(line) == list:
for l in line:
plt.axhline(y=l, color="red", linestyle="--")
else:
plt.axhline(y=line, color="red", linestyle="--")
plt.legend()
plt.title(column2)
plt.tight_layout()
plt.show()
# 表示例
plot_category_category(df, "Pclass", "Sex", scale=0.7, line=100)
- 元のデータとの割合線も表示するバージョン
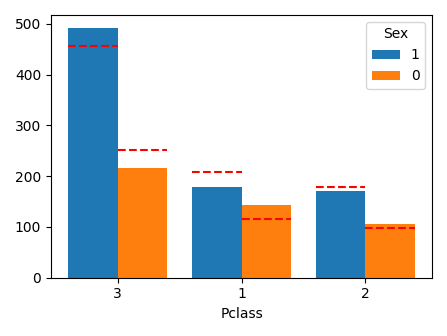
def plot_category_category(df, column1, column2, scale=1.0):
plt.figure(figsize=(6.4*scale, 4.8*scale))
# 欠損値が入っているデータは除外する
df = df.dropna(subset=[column1, column2]).copy()
# 型を変換
df[column1] = df[column1].astype(str)
df[column2] = df[column2].astype(str)
# マージンを設定
margin = 0.2
totoal_width = 1 - margin
# hue_data
hue_counts = df[column2].value_counts()
hue_keys = hue_counts.keys()
# clumn_data
column_keys = df[column1].value_counts().keys()
hue_column_counts = {}
for k2 in hue_keys:
hue_column_counts[k2] = []
for k in column_keys:
c = df[df[column1]==k][column2].value_counts()
# hue key毎に分けておく
for k2 in hue_keys:
if k2 in c:
hue_column_counts[k2].append(c[k2])
else:
hue_column_counts[k2].append(0)
column_counts = []
for k in column_keys:
c = len(df[df[column1]==k])
column_counts.append(c)
# barの座標を計算
bar_count = len(hue_keys)
bar_width = totoal_width/bar_count
x_bar_pos = np.asarray(range(bar_count)) * bar_width
for i, k in enumerate(hue_keys):
counts = hue_column_counts[k]
# barを表示
x_pos = np.asarray(range(len(counts)))
x_pos = x_pos + x_bar_pos[i] - (bar_count * bar_width) / 2 + bar_width / 2
plt.bar(x_pos, counts, width=bar_width, label=hue_keys[i])
# 線を表示
p_all = (hue_counts[k] / hue_counts.sum())
c = np.asarray(column_counts) * p_all
plt.hlines(c, xmin=x_pos - bar_width/2, xmax=x_pos + bar_width/2, colors='r', linestyles='dashed')
plt.xticks(range(len(column_keys)), column_keys)
plt.xlabel(column1)
plt.legend(title=column2)
plt.tight_layout()
plt.show()
# 表示例
plot_category_category(df, "Pclass", "Sex", scale=0.7)
print表示
- 集計
print(pd.crosstab(df["Pclass"], df["Sex"]))
Sex female male
Pclass
1 144 179
2 106 171
3 216 493
- 各Sexにおける、Pclassの比率
print(pd.crosstab(df["Pclass"], df["Sex"], normalize='columns'))
Sex female male
Pclass
1 0.309013 0.212337
2 0.227468 0.202847
3 0.463519 0.584816
Sex female male
- 各Pclassにおける、Sexの割合
print(pd.crosstab(df["Pclass"], df["Sex"], normalize='index'))
Sex female male
Pclass
1 0.445820 0.554180
2 0.382671 0.617329
3 0.304654 0.695346
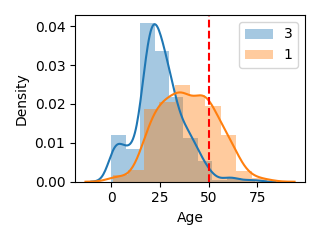
カテゴリデータに対して数値データを比較したい(2変数)
import math
def plot_category_float(
df,
column1,
column2,
bins=10,
exclude=[],
scale=1.0,
line=None,
):
plt.figure(figsize=(6.4*scale, 4.8*scale))
for uniq in df[column1].unique():
if uniq is np.nan:
continue
if math.isnan(uniq):
continue
if uniq in exclude:
continue
# 表示メイン
sns.distplot(df[df[column1]==uniq][column2], kde=True, rug=False, bins=bins, label=uniq)
if line is not None:
if type(line) == list:
for l in line:
plt.axvline(x=l, color="red", linestyle="--")
else:
plt.axvline(x=line, color="red", linestyle="--")
plt.legend()
plt.tight_layout()
plt.show()
# 表示例
plot_category_float(df, "Pclass", "Age", exclude=[2], scale=0.5, line=50)
※ascendingはNoneの場合ソートしません。
指定する場合はpandasのsort_valuesの引数と同じです。
※excludeを指定すると任意のカテゴリデータを非表示にできます。
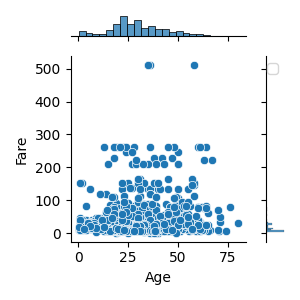
数値データに対して数値データを比較したい(2変数)
def plot_float_float(
df,
column1,
column2,
hue=None,
scale=1.0,
):
# 表示メイン
sns.jointplot(column1, column2, data=df, hue=hue, palette='Set2', size=6*scale)
plt.legend()
plt.tight_layout()
plt.show()
# 表示例
plot_float_float(df, "Age", "Fare", "Pclass", scale=0.5)
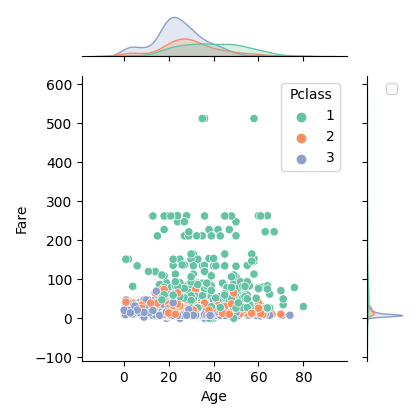
数値データと数値データに対してカテゴリデータを比較したい(3変数)
plot_float_floatのhueを指定したバージョンです。
def plot_float_float_category(df, column1, column2, column3, scale=1.0,):
plot_float_float(df, column1, column2, column3, scale)
# 表示例
plot_float_float_category(df, "Age", "Fare", "Pclass", scale=0.7)
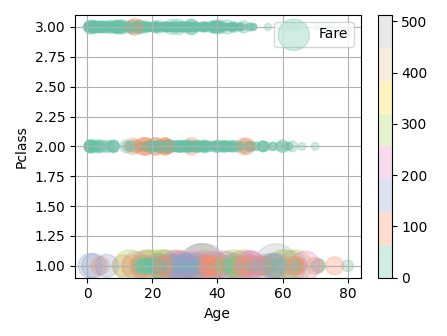
数値データと数値データに対して数値データを比較したい(3変数)
def plot_float_float_float(
df,
column1,
column2,
column3,
scale=1.0,
):
_ds3 = (df[column3] - df[column3].min()) / (df[column3].max() - df[column3].min())
_ds3 = 10 + _ds3*1000
fig = plt.figure(figsize=(6.4*scale, 4.8*scale))
mappable = plt.scatter(df[column1], df[column2], s=_ds3, c=df[column3], cmap="Set2", alpha=0.3, label=column3)
fig.colorbar(mappable)
plt.xlabel(column1)
plt.ylabel(column2)
plt.grid(True)
plt.legend()
plt.tight_layout()
plt.show()
# 表示例
plot_float_float_float(df, "Age", "Pclass", "Fare", scale=0.7)
※表示例はPclassなので小数データではないですが…
※cmapの種類についてはここを参考
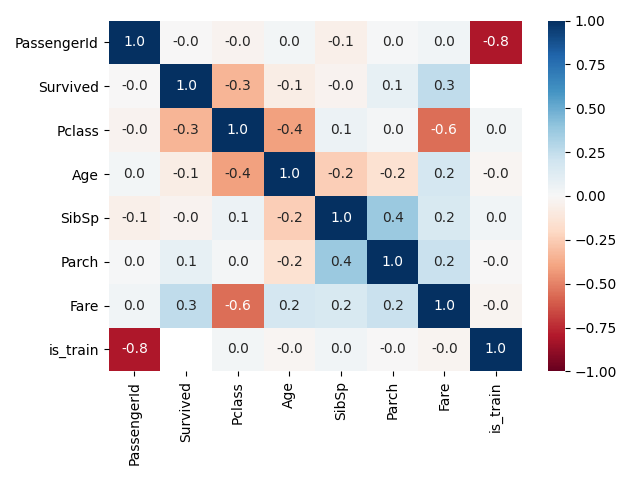
変数同士の相関係数をみたい
- ヒートマップの表示
def plot_corr(df, columns, scale=1.0):
plt.figure(figsize=(6.4*scale, 4.8*scale))
# 表示メイン
sns.heatmap(df[columns].corr(), annot=True, vmax=1, vmin=-1, fmt='.1f', cmap='RdBu')
plt.tight_layout()
plt.show()
# 表示例
plot_corr(df, df.columns)
- ある変数に対して相関係数が高い変数のみ表示(1対多)
def print_corr(df, columns, threshold=0.5):
print("--- 相関係数(threshold: {})".format(threshold))
corr = df[columns].corr()
count = 0
for i in range(len(corr.columns)):
for j in range(i+1, len(corr.columns)):
val = corr.iloc[i, j]
if abs(val) > threshold:
print("{} {}: {:.2f}".format(corr.columns[i], corr.columns[j], val))
count += 1
if count == 0:
print("empty")
# 表示例
print_corr(df, df.columns, threshold=0.4)
--- 相関係数(threshold: 0.4)
PassengerId is_train: -0.81
Pclass Age: -0.41
Pclass Fare: -0.56
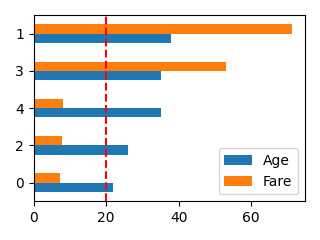
ある配列データを横棒グラフで表示して比較したい
def plot_bar(df, columns, ascending=None, scale=1.0, line=None):
if ascending is None:
df.plot.barh(y=columns, figsize=(6.4*scale, 4.8*scale))
else:
df.sort_values(columns, ascending=ascending).plot.barh(y=columns, figsize=(6.4*scale, 4.8*scale))
if line is not None:
if type(line) == list:
for l in line:
plt.axvline(x=l, color="red", linestyle="--")
else:
plt.axvline(x=line, color="red", linestyle="--")
plt.tight_layout()
plt.show()
# 表示例
plot_bar(df[:5], ["Age", "Fare"], ascending=True, scale=0.5, line=20)
※ascendingがNoneの場合は並べ替えしません。
それ以外の場合はpandasのsort_valuesの引数と同じ動作です
3.データ加工
欠損値を補完
df["Age_org"] = df["Age"] # 後で使うので欠損値がある状態も保存
df["Age_na"] = df["Age"].isnull()
df["Age"].fillna(df["Age_org"].median(), inplace=True)
df["Embarked"].fillna("S", inplace=True)
df["Fare"].fillna(df['Fare'].median(), inplace=True)
Label Encording
from sklearn.preprocessing import LabelEncoder
df["Sex"] = LabelEncoder().fit_transform(df['Sex'])
df["Embarked"] = LabelEncoder().fit_transform(df['Embarked'])
ダミー化(One-hotエンコーディング)
pandasのget_dummiesで簡単にできますが、カラム名も欲しい場合のものを作成しています。
def dummy(df, column):
c_arr = []
for c in df[column].value_counts().keys():
name = column + "_" + str(c)
c_arr.append(name)
df[name] = np.asarray(df[column] == c).astype(int)
return df, c_arr
4.目的変数に対する各説明変数の影響度を確認したい
確認に使っているカラムは以下です。
(Familyも何となく追加)
df["Family"] = df["SibSp"] + df["Parch"]
select_columns = [
"Pclass",
"Sex",
"Age",
"SibSp",
"Parch",
"Family",
"Fare",
"Embarked",
]
分類
- ロジスティック回帰の係数
- ランダムフォレストにおける各変数の重要度
- XGBoostにおける各変数の重要度
- LightGBMにおける各変数の重要度
import sklearn.linear_model
import xgboost as xgb
import lightgbm as lgb
def create_feature_effect_classifier(df, select_columns, target_column):
# select_columnsにtarget_columnが入っていれば削除
select_columns = list(select_columns)
if target_column in select_columns:
select_columns.remove(target_column)
x = df[df[target_column].notnull()][select_columns]
y = df[df[target_column].notnull()][target_column]
# 説明変数に欠損値があると判定できない手法があるので
exists_missing_value = (np.sum(x.isnull().sum()) > 0)
# 結果用
df_result = pd.DataFrame(select_columns, columns=["name"])
#----------------
# ロジスティック回帰
#----------------
if not exists_missing_value:
model = sklearn.linear_model.LogisticRegression(random_state=1234)
model.fit(x, y)
if len(model.coef_) == 1:
# 2クラス分類
df_result["LogisticRegression"] = model.coef_[0]
else:
# 多クラスの場合は各クラスの係数の平均を出す
df_result["LogisticRegression"] = np.mean(model.coef_, axis=0)
#----------------
# RandomForest
#----------------
if not exists_missing_value:
model = sklearn.ensemble.RandomForestClassifier(random_state=1234)
model.fit(x, y)
df_result["RandomForest"] = model.feature_importances_
#----------------
# XGB
#----------------
model = xgb.XGBClassifier(eval_metric="logloss", use_label_encoder=False, random_state=1234)
model.fit(x, y)
df_result["XGB"] = model.feature_importances_
#----------------
# LGBM
#----------------
model = lgb.LGBMClassifier(random_state=1234)
model.fit(x, y)
df_result["LGBM"] = model.feature_importances_
df_result.set_index("name", inplace=True)
return df_result
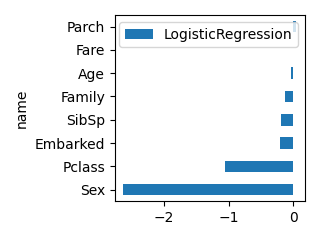
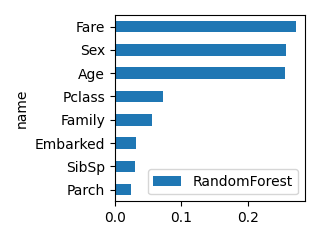
# 実行例
df_feature = create_feature_effect_classifier(df, select_columns, "Survived")
print(df_feature)
plot_bar(df_feature, "LogisticRegression", ascending=True, scale=0.5)
plot_bar(df_feature, "RandomForest", ascending=True, scale=0.5)
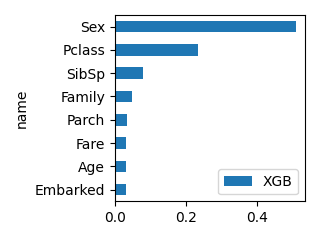
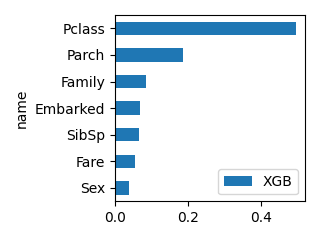
plot_bar(df_feature, "XGB", ascending=True, scale=0.5)
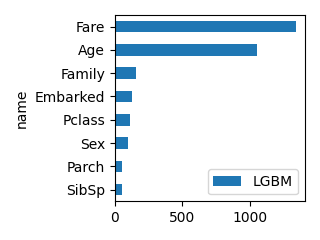
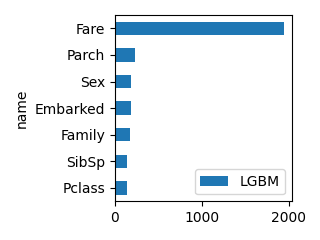
plot_bar(df_feature, "LGBM", ascending=True, scale=0.5)
LogisticRegression RandomForest XGB LGBM
name
Pclass -1.050753 0.072651 0.233783 116
Sex -2.631111 0.257130 0.509833 97
Age -0.038077 0.255461 0.032465 1055
SibSp -0.182407 0.030305 0.078906 51
Parch 0.047843 0.024342 0.034393 56
Family -0.134563 0.055398 0.047479 154
Fare 0.002171 0.272152 0.032554 1344
Embarked -0.213182 0.032561 0.030586 125
回帰
- 重回帰分析の決定係数(とP値)
- ランダムフォレストにおける各変数の重要度
- XGBoostにおける各変数の重要度
- LightGBMにおける各変数の重要度
import statsmodels.api as sm
import sklearn.ensemble
import xgboost as xgb
import lightgbm as lgb
def create_feature_effect_regressor(df, select_columns, target_column):
# select_columnsにtarget_columnが入っていれば削除
select_columns = list(select_columns)
if target_column in select_columns:
select_columns.remove(target_column)
x = df[df[target_column].notnull()][select_columns]
y = df[df[target_column].notnull()][target_column]
# 説明変数に欠損値があると判定できない手法があるので
exists_missing_value = (np.sum(x.isnull().sum()) > 0)
# 結果用
df_result = pd.DataFrame(select_columns, columns=["name"])
#----------------
# 重回帰分析
#----------------
if not exists_missing_value:
model = sm.OLS(y, sm.add_constant(x))
fitted = model.fit()
#print(fitted.summary())
if len(fitted.params) == len(select_columns):
df_result["OLS_val"] = list(fitted.params)
df_result["OLS_p"] = list(fitted.pvalues)
elif len(fitted.params) == len(select_columns)+1:
df_result["OLS_val"] = list(fitted.params)[1:]
df_result["OLS_p"] = list(fitted.pvalues)[1:]
df_result["OLS_p"] = np.round(df_result["OLS_p"], 3)
#----------------
# RandomForest
#----------------
if not exists_missing_value:
model = sklearn.ensemble.RandomForestRegressor(random_state=1234)
model.fit(x, y)
df_result["RandomForest"] = model.feature_importances_
#----------------
# XGB
#----------------
model = xgb.XGBRegressor(eval_metric="logloss", use_label_encoder=False, random_state=1234)
model.fit(x, y)
df_result["XGB"] = model.feature_importances_
#----------------
# LGBM
#----------------
model = lgb.LGBMRegressor(random_state=1234)
model.fit(x, y)
df_result["LGBM"] = model.feature_importances_
df_result.set_index("name", inplace=True)
return df_result
# 表示例
df_feature = create_feature_effect_regressor(df, select_columns, "Age")
print(df_feature)
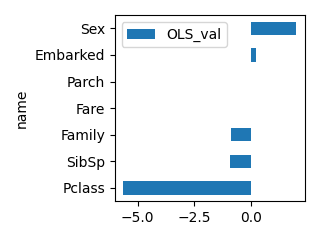
plot_bar(df_feature, "OLS_val", ascending=True, scale=0.5)
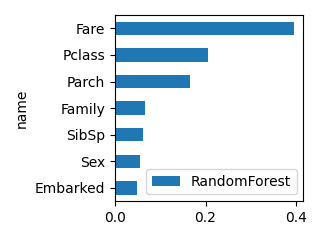
plot_bar(df_feature, "RandomForest", ascending=True, scale=0.5)
plot_bar(df_feature, "XGB", ascending=True, scale=0.5)
plot_bar(df_feature, "LGBM", ascending=True, scale=0.5)
OLS_val OLS_p RandomForest XGB LGBM
name
Pclass -5.622647 0.000 0.205440 0.496402 140
Sex 2.020035 0.004 0.054703 0.038413 190
SibSp -0.910016 0.003 0.062044 0.066298 141
Parch 0.036394 0.913 0.164997 0.187804 234
Family -0.873621 0.000 0.066830 0.086484 171
Fare 0.006825 0.400 0.395958 0.056573 1940
Embarked 0.244200 0.556 0.050028 0.068026 184
P値(OLD_p)が0.05以上の場合は有意性がありません。(決定係数(OLS_val)が0の可能性があります)
今回ですと、Parch,Fare,Embarked は有意性がないので説明変数としては採用しないほうが良いかもしれません。
決定木
import sklearn.tree
def plot_decision_tree(df, columns, target_column,
model_type, # "Classifier" or "Regressor"
scale=1.0,
max_depth=None, # 表示する木の深さ
fontsize=None, # 文字の大きさ
):
x = df[df[target_column].notnull()][columns]
y = df[df[target_column].notnull()][target_column]
if model_type == "Classifier":
clf = sklearn.tree.DecisionTreeClassifier(random_state=1234)
elif model_type == "Regressor":
clf = sklearn.tree.DecisionTreeRegressor(random_state=1234)
else:
raise ValueError()
clf = clf.fit(x, y)
plt.figure(figsize=(6.4*scale, 4.8*scale))
sklearn.tree.plot_tree(clf, feature_names=columns,
max_depth=max_depth,
fontsize=fontsize,
proportion=False, # Trueだと割合で表示
filled=True, # 純度に応じて色がつく
)
plt.show()
# 表示例
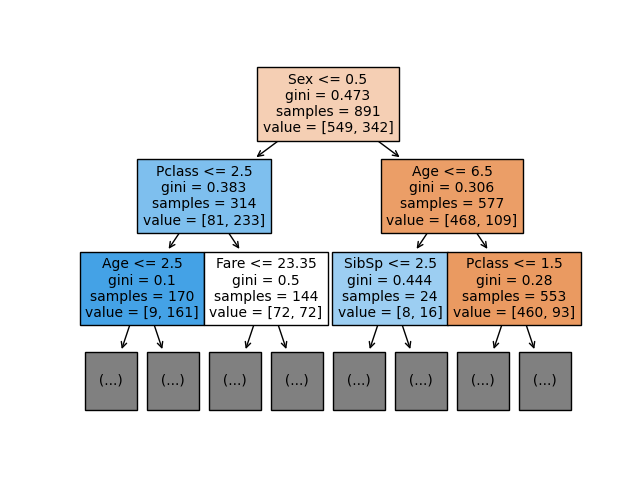
plot_decision_tree(df, select_columns, "Survived", "Classifier",
scale=1, max_depth=2, fontsize=10)
5.Validation
K-分割交差検証
層状K-分割交差検証の実装例です。
def validation_classifier(
df, # 全データ(DataFrame)
select_columns, # 説明変数のリスト
target_column, # 目的変数
model_cls, # 使うモデル
model_params, # モデルのパラメータ
):
# select_columnsにtarget_columnが入っていれば削除
select_columns = list(select_columns)
if target_column in select_columns:
select_columns.remove(target_column)
# 学習用データを作成
df_train = df[df[target_column].notnull()].copy()
# 交差検証
metrics = []
#kf = sklearn.model_selection.KFold(n_splits=3, shuffle=True, random_state=1234) # KFoldにしたい場合
kf = sklearn.model_selection.StratifiedKFold(n_splits=3, shuffle=True, random_state=1234)
for train_idx, test_idx in kf.split(df_train, y=df_train[target_column]):
df_train_sub = df_train.iloc[train_idx]
df_test_sub = df_train.iloc[test_idx]
x_train = df_train_sub[select_columns]
y_train = df_train_sub[target_column]
x_test = df_test_sub[select_columns]
y_test = df_test_sub[target_column]
model = model_cls(**model_params)
model.fit(x_train, y_train)
# 正解率
y_pred = model.predict(x_test)
metric = sklearn.metrics.accuracy_score(y_test, y_pred)
# logloss
#y_pred = model.predict_proba(x_test)
#metric = sklearn.metrics.log_loss(y_test, y_pred[:,1])
metrics.append(metric)
# 交差検証の結果は平均を採用
metrics_mean = np.mean(metrics, axis=0)
return metrics_mean
# 実行例
metric = validation_classifier(df, select_columns, "Survived",
lgb.LGBMClassifier, {"random_state": 1234})
print(metric)
0.8103254769921437
Adversarial Validation
学習データとテストデータの分布を確認し、分布が偏っている場合にテストデータに近いデータを検証に使う手法です。
モデルはLGBMで決め打ちしています。
import sklearn.model_selection
import sklearn.metrics
import lightgbm as lgb
def adversarial_validation_check(
df, # 全データ(DataFrame)
select_columns, # 説明変数のリスト
target_column, # 目的変数
result_column_name="", # 結果保存用のカラム名
ebable_plot=True,
):
# モデル
model = lgb.LGBMClassifier(**{"random_state": 1234})
# select_columnsにtarget_columnが入っていれば削除
select_columns = list(select_columns)
if target_column in select_columns:
select_columns.remove(target_column)
# 学習用データを作成
df_train = df.copy()
# 目的変数を作成
df_train["_target"] = df_train[target_column].apply(lambda x: np.isnan(x)).astype(int)
# 学習データとテストデータに分割
df_train_sub, df_true_sub = sklearn.model_selection.train_test_split(
df_train, test_size=0.3, stratify=df_train["_target"], random_state=1234)
x_train = df_train_sub[select_columns]
y_train = df_train_sub["_target"]
x_true = df_true_sub[select_columns]
y_true = df_true_sub["_target"]
# 学習
model.fit(x_train, y_train, eval_set=[(x_train, y_train), (x_true, y_true)], eval_names=['train', 'valid'], eval_metric='auc', verbose=0)
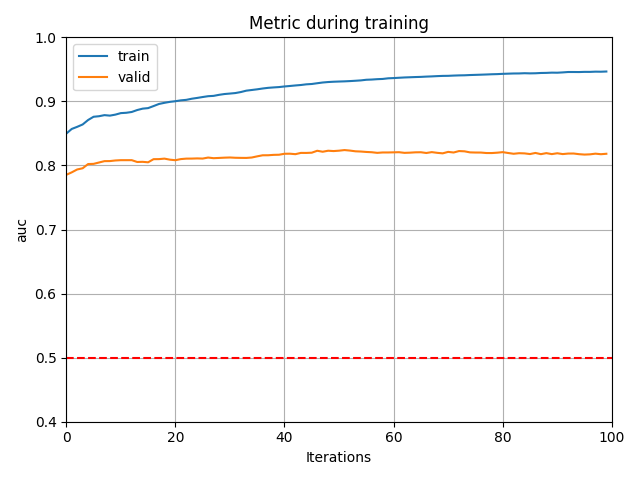
if ebable_plot:
# 学習結果をplot
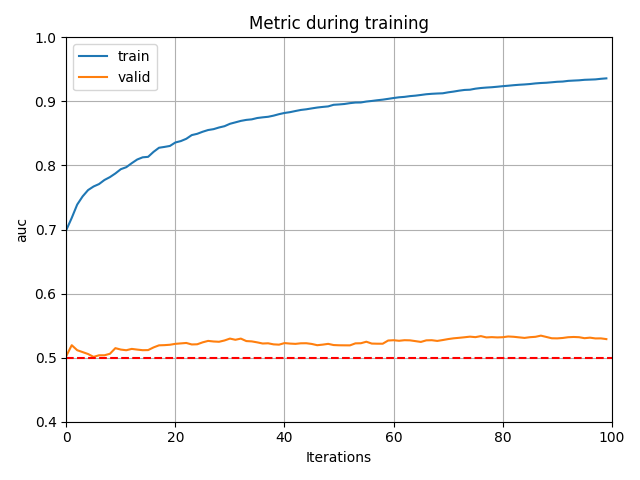
lgb.plot_metric(model.evals_result_, metric='auc')
plt.axhline(y=0.5, color="red", linestyle="--")
plt.ylim(0.4, 1.0)
plt.tight_layout()
plt.show()
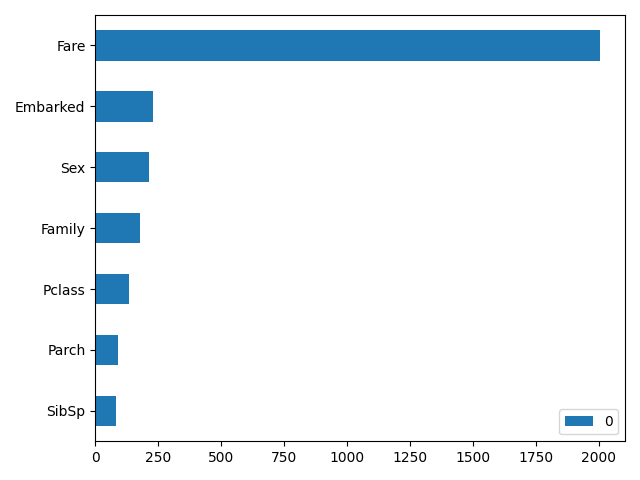
# 分布に影響が大きい変数順に並べて表示する
pd.DataFrame(model.feature_importances_, index=select_columns
).sort_values(0, ascending=True).plot.barh()
plt.tight_layout()
plt.show()
# 結果を新しい特徴量として追加
if result_column_name != "":
y_pred = model.predict_proba(x_true)
df[result_column_name] = pd.Series(y_pred[:,1])
# auc値を返す
return sklearn.metrics.roc_auc_score(y_true, y_pred[:,1])
# 実行例
adversarial_validation_check(df, select_columns, "Survived")
adversarial_validation_check(df, select_columns, "Age_org", "train_ratio_Age")
- Survivedの結果
valid の値が0.5付近なので、学習データとテストデータを区別できない=学習データとテストデータの分布が同じです。
この場合はAdversarial Validationを使う必要はありません。
- Ageの結果
Ageは欠損値のないデータ群と欠損値のあるデータ群を比較しています。
validが約0.8とかなり高い確率で予測できています。
Ageの欠損値がある場合とない場合で分布に偏りがある事が分かります。
また、変数の重要度を見ることで分布の偏りの原因になっている変数が分かります。
(今回はFareが一番影響しているようですね)
これを用いて検証するコード例です。
上記の予測結果に対して、テストデータである確率が高いデータを検証データに、そうじゃないデータは学習データにして検証を行います。
def adversarial_validation_Age(
df, # 全データ(DataFrame)
select_columns, # 説明変数のリスト
model_cls, # 使うモデル
model_params, # モデルのパラメータ
):
target_column = "Age_org"
# select_columnsにtarget_columnが入っていれば削除
select_columns = list(select_columns)
if target_column in select_columns:
select_columns.remove(target_column)
# 学習用データを作成
df_train = df[df[target_column].notnull()].copy()
# データをtrain_ratioに従って分割
test_size = 0.25
test_num = int(len(df_train) * test_size)
df_train = df_train.sort_values("train_ratio_Age")
df_train_sub = df_train[test_num:]
df_test_sub = df_train[:test_num]
x_train = df_train_sub[select_columns]
y_train = df_train_sub[target_column]
x_test = df_test_sub[select_columns]
y_test = df_test_sub[target_column]
# 学習
model = model_cls(**model_params)
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
# 評価
metric = np.sqrt(sklearn.metrics.mean_squared_error(y_test, y_pred))
return metric
metric = adversarial_validation_Age(df, select_columns, lgb.LGBMRegressor, {"random_state": 1234})
print(print)
12.119246573863585
6.特徴量抽出
参考:特徴量選択のまとめ
全く関係ない特徴量を減らしたい
- 分散が0(すべて同じ値)データを削除
- 全く同じ特徴量のカラムを削除
import sklearn.feature_selection
def feature_selection_no_related(
df,
columns,
enable_print=False
):
prev_columns = columns
prev_len = len(prev_columns)
#-------------------------------
# 分散が0(すべて同じ値)のカラムは削除
#-------------------------------
vt = sklearn.feature_selection.VarianceThreshold(threshold=0)
vt.fit(df[prev_columns])
inc_columns = []
exc_columns = []
for i, flag in enumerate(vt.get_support()):
if flag:
inc_columns.append(prev_columns[i])
else:
exc_columns.append(prev_columns[i])
if enable_print:
print("var0 {} -> {} exc: {}".format(prev_len, len(inc_columns), exc_columns))
#-------------------------------
# 全く同じ特徴量のカラムは削除
#-------------------------------
prev_columns = inc_columns
prev_len = len(prev_columns)
inc_columns = []
exc_columns = []
for c, flag in df[prev_columns].T.duplicated().items():
if not flag:
inc_columns.append(c)
else:
exc_columns.append(c)
if enable_print:
print("dup {} -> {} exc: {}".format(prev_len, len(inc_columns), exc_columns))
return inc_columns
# 実行例
df["A"] = 1
df["B"] = df["Sex"]
new_columns = feature_reduction_no_related(df,
select_columns + ["A", "B"], enable_print=True)
print(new_columns)
var0 10 -> 9 exc: ['A']
dup 9 -> 8 exc: ['B']
['Pclass', 'Sex', 'Age', 'SibSp', 'Parch', 'Family', 'Fare', 'Embarked']
多分関係ない特徴量を減らしたい
相関係数
相関係数は0なら相関なし、1または-1なら正か負の強い相関があります。
0に近い特徴量を減らします。
def feature_selection_corr(
df,
columns,
target_columns,
threshold,
enable_print=False
):
# target_columnをいれる
prev_columns = list(set(list(columns) + [target_columns]))
prev_len = len(prev_columns)
# 相関係数
df_corr = df[prev_columns].corr()
df_corr.fillna(0, inplace=True)
# 閾値以下は除外
inc_columns = []
exc_columns = []
for c in df_corr.columns:
val = df_corr[target_columns][c]
if abs(val) < threshold:
exc_columns.append([c, val])
else:
inc_columns.append(c)
if enable_print:
print("corr {} -> {} exc: {}".format(prev_len, len(inc_columns), exc_columns))
return inc_columns
# 実行例
new_columns = feature_selection_corr(df, select_columns,
"Survived", 0.1, enable_print=True)
print(new_columns)
corr 9 -> 5 exc: [['SibSp', -0.03532249888573556], ['Parch', 0.08162940708348335], ['Age', -0.07722109457217759], ['Family', 0.016638989282745195]]
['Embarked', 'Fare', 'Sex', 'Survived', 'Pclass']
関係のある特徴量のみを抽出したい
評価の上がる特徴量を追加(または削除)していく
全説明変数を見て一番評価が上がる変数を追加または削除していく手法です。
Fowardは説明変数がない状態から評価が上がる変数を1つずつ追加していく方法で、
Backwardは全説明変数から評価が下がる変数を1つずつ減らしていく方法です。
説明変数の数に対して最大O(N^2)時間がかかると思います。
sklearnのライブラリを使用する場合
import sklearn.feature_selection
def feature_selection_SFS(
df,
columns,
target_columns,
model_cls, # 使うモデル
model_params, # モデルのパラメータ
direction, # "forward"で追加していく、"backward"で削除していく
scoring, # 評価方法
):
x = df[df[target_columns].notnull()][columns]
y = df[df[target_columns].notnull()][target_columns]
rfe = sklearn.feature_selection.SequentialFeatureSelector(
model_cls(**model_params),
direction=direction,
scoring=scoring
)
rfe.fit(x, y)
inc_columns = []
exc_columns = []
for i, flag in enumerate(rfe.get_support()):
if flag:
inc_columns.append(columns[i])
else:
exc_columns.append(columns[i])
return inc_columns, exc_columns
# 実行例
inc_columns, exc_columns = feature_selection_SFS(
df, select_columns, "Survived",
lgb.LGBMClassifier, {"random_state": 1234},
direction="forward",
scoring="accuracy")
print("inc", inc_columns)
print("exc", exc_columns)
inc ['Pclass', 'Sex', 'Age', 'Fare']
exc ['SibSp', 'Parch', 'Family', 'Embarked']
手動でコードを書く場合(増やす)
def feature_selection_forward(df, columns, target_column, lookahead_times=5):
inc_columns = []
best_columns = []
best_metric = 0
lookahead_count = 0
for _ in tqdm(range(len(columns))):
valid_list = []
for c in columns:
if c in inc_columns:
continue
_test_columns = inc_columns[:]
_test_columns.append(c)
metric = validation_classifier(df, _test_columns, target_column, lgb.LGBMClassifier, {"random_state": 1234})
valid_list.append([metric, c])
valid_list.sort()
best_valid = valid_list[-1]
inc_columns.append(best_valid[1])
print(best_valid)
if best_valid[0] >= best_metric:
best_metric = best_valid[0]
best_columns = inc_columns[:]
lookahead_count = 0
else:
lookahead_count += 1
if lookahead_count >= lookahead_times:
break
return best_columns
手動でコードを書く場合(減らす)
def feature_selection_backword(df, columns, target_column, lookahead_times=5):
inc_columns = [:]
best_columns = [:]
best_metric = 0
lookahead_count = 0
for _ in tqdm(range(len(columns)-1)):
valid_list = []
for c in columns:
if c not in inc_columns:
continue
_test_columns = inc_columns[:]
_test_columns.remove(c)
metric = validation_classifier(df, _test_columns, target_column, lgb.LGBMClassifier, {"random_state": 1234})
valid_list.append([metric, c])
valid_list.sort()
best_valid = valid_list[-1]
inc_columns.remove(best_valid[1])
print(best_valid)
if best_valid[0] >= best_metric:
best_metric = best_valid[0]
best_columns = inc_columns[:]
lookahead_count = 0
else:
lookahead_count += 1
if lookahead_count >= lookahead_times:
break
return best_columns
手動でコードを書く場合(総当たり)
import itertools
def feature_selection_all(df, columns, target_column):
columns_list = []
for i in range(1, len(columns)):
for c in itertools.combinations(columns, i):
columns_list.append(c)
arr = []
for cin tqdm(columns_list):
m = train_cv(df, c, target_column, n_splits=5)
arr.append([m, c])
arr.sort()
pprint(arr[-10:])
return arr[-1][1] # best_columns
RFECV
RFECVは簡単に言うとモデルを作成し、重要度が低い変数を除外するという特徴量選択方法らしいです。
その方法から、coef_またはfeature_importances_が作られるモデルしか適用できません。
参考:【Kaggle】タイタニックの振り返り#3 RFECVで特徴選択
import sklearn.feature_selection
def feature_selection_RFECV(
df,
columns,
target_columns,
model_cls, # 使うモデル
model_params, # モデルのパラメータ
scoring, # 評価方法
):
x = df[df[target_columns].notnull()][columns]
y = df[df[target_columns].notnull()][target_columns]
rfe = sklearn.feature_selection.RFECV(model_cls(**model_params), scoring=scoring)
rfe.fit(x, y)
inc_columns = []
exc_columns = []
for i, flag in enumerate(rfe.get_support()):
if flag:
inc_columns.append(columns[i])
else:
exc_columns.append(columns[i])
return inc_columns, exc_columns
# 実行例
inc_columns, exc_columns = feature_selection_RFECV(
df, select_columns, "Survived",
lgb.LGBMClassifier, {"random_state": 1234},
scoring="accuracy")
print("inc", inc_columns)
print("exc", exc_columns)
inc ['Pclass', 'Sex', 'Age', 'SibSp', 'Family', 'Fare', 'Embarked']
exc ['Parch']
7.モデル
モデル一覧
分類モデル
import sklearn.ensemble
import sklearn.gaussian_process
import sklearn.linear_model
import sklearn.naive_bayes
import sklearn.neighbors
import sklearn.tree
import sklearn.svm
import sklearn.discriminant_analysis
import sklearn.neural_network
import xgboost as xgb
import lightgbm as lgb
import catboost
def get_models_classifier(random_seed):
models = [
#Ensemble Methods
[sklearn.ensemble.AdaBoostClassifier, {"random_state": random_seed}],
[sklearn.ensemble.BaggingClassifier, {"random_state": random_seed}],
[sklearn.ensemble.ExtraTreesClassifier, {"random_state": random_seed}],
[sklearn.ensemble.GradientBoostingClassifier, {"random_state": random_seed}],
[sklearn.ensemble.RandomForestClassifier, {"random_state": random_seed}],
#Gaussian Processes
[sklearn.gaussian_process.GaussianProcessClassifier, {"random_state": random_seed}],
#GLM
[sklearn.linear_model.LogisticRegression, {"random_state": random_seed}, "score"],
[sklearn.linear_model.RidgeClassifier, {}],
#Navies Bayes
[sklearn.naive_bayes.BernoulliNB, {}],
[sklearn.naive_bayes.GaussianNB, {}],
#Nearest Neighbor
[sklearn.neighbors.KNeighborsClassifier, {}],
#Trees
[sklearn.tree.DecisionTreeClassifier, {"random_state": random_seed}],
[sklearn.tree.ExtraTreeClassifier, {"random_state": random_seed}],
# SVM
[sklearn.svm.SVC, {}],
[sklearn.svm.NuSVC, {}],
[sklearn.svm.LinearSVC, {}],
# NN
[sklearn.neural_network.MLPClassifier, {"random_state": random_seed}],
#xgboost
[xgb.XGBClassifier, {"eval_metric": "logloss", "use_label_encoder": False, "random_state": random_seed}],
# light gbm
[lgb.LGBMClassifier, {"random_state": random_seed}],
# catboost
[catboost.CatBoostClassifier, {"verbose":0, "random_seed": 1234}],
]
return models
models = get_models_classifier(1234)
回帰モデル
import sklearn.ensemble
import sklearn.linear_model
import sklearn.naive_bayes
import sklearn.neighbors
import sklearn.tree
import sklearn.svm
import sklearn.neural_network
import xgboost as xgb
import lightgbm as lgb
import catboost
def get_models_regressor(random_seed):
models = [
#Ensemble Methods
[sklearn.ensemble.AdaBoostRegressor, {"random_state": random_seed}],
[sklearn.ensemble.BaggingRegressor, {"random_state": random_seed}],
[sklearn.ensemble.ExtraTreesRegressor, {"random_state": random_seed}],
[sklearn.ensemble.GradientBoostingRegressor, {"random_state": random_seed}],
[sklearn.ensemble.RandomForestRegressor, {"random_state": random_seed}],
# GLM
[sklearn.linear_model.Lasso, {"random_state": random_seed}],
[sklearn.linear_model.Ridge, {"random_state": random_seed}],
[sklearn.linear_model.ElasticNet, {"random_state": random_seed}],
#Nearest Neighbor
[sklearn.neighbors.KNeighborsRegressor, {}],
#Trees
[sklearn.tree.DecisionTreeRegressor, {"random_state": random_seed}],
[sklearn.tree.ExtraTreeRegressor, {"random_state": random_seed}],
# SVM
[sklearn.svm.SVR, {}],
[sklearn.svm.NuSVR, {}],
[sklearn.svm.LinearSVR, {}],
# NN
[sklearn.neural_network.MLPRegressor, {"random_state": random_seed}],
#xgboost
[xgb.XGBRegressor, {"eval_metric": "logloss", "use_label_encoder": False, "objective": 'reg:squarederror', "random_state": random_seed}],
# light gbm
[lgb.LGBMRegressor, {"random_state": random_seed}],
# catboost
[catboost.CatBoostRegressor, {"verbose":0, "random_seed": 1234}]
]
return models
models = get_models_regressor(1234)
モデルの比較コード例
from tqdm import tqdm
df_model = pd.DataFrame(columns=["metric"])
for model in tqdm(get_models_classifier(1234)):
try:
#--- モデルを評価するコードを書く
metric = validation_classifier(df, select_columns, "Survived", model[0], model[1])
#---
df_model.loc[model[0].__name__] = metric
except:
import traceback
print(model[0].__name__)
print(traceback.print_exc())
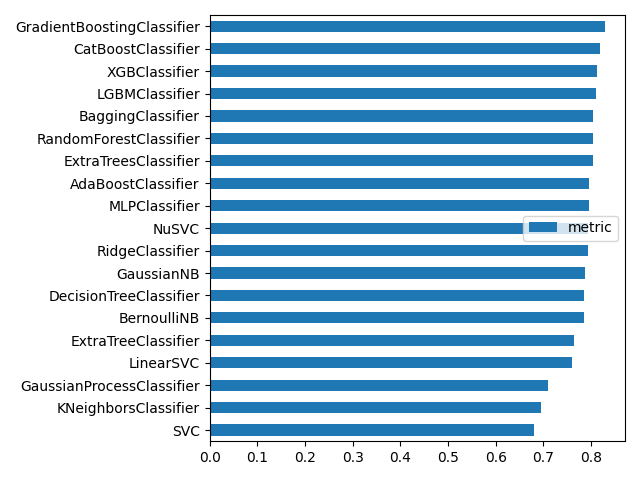
print(df_model.sort_values("metric"))
plot_bar(df_model, "metric", ascending=True)
metric
SVC 0.680135
KNeighborsClassifier 0.694725
GaussianProcessClassifier 0.710438
LinearSVC 0.759820
ExtraTreeClassifier 0.764310
BernoulliNB 0.784512
DecisionTreeClassifier 0.784512
GaussianNB 0.786756
RidgeClassifier 0.792368
NuSVC 0.793490
MLPClassifier 0.794613
AdaBoostClassifier 0.795735
ExtraTreesClassifier 0.804714
RandomForestClassifier 0.804714
BaggingClassifier 0.804714
LGBMClassifier 0.810325
XGBClassifier 0.812570
CatBoostClassifier 0.819304
GradientBoostingClassifier 0.829405
ハイパーパラメータの調整(optuna)
モデルパラメータ
**クリックで展開**
def create_optuna_params(trial, model_cls, random_seed):
def not_support_pattern(params):
return False
#-----------------------------------------------------------
# Classifier
#-----------------------------------------------------------
#Ensemble Methods
if model_cls == sklearn.ensemble.AdaBoostClassifier:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=50
"learning_rate": trial.suggest_uniform('learning_rate', 0, 1), # default=1
"algorithm": trial.suggest_categorical('algorithm', ["SAMME", "SAMME.R"]), # default=SAMME.R
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.BaggingClassifier:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 500), # default=10
#"max_samples": trial.suggest_uniform('max_samples', 0, 1), # default=1
"max_features": trial.suggest_uniform('max_features', 0, 1), # default=1
"bootstrap": trial.suggest_categorical('bootstrap', [False, True]), # default=True
"bootstrap_features": trial.suggest_categorical('bootstrap_features', [False, True]), # default=False
# oob_score
# warm_start
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.ExtraTreesClassifier:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 100), # default=100
"criterion": trial.suggest_categorical('criterion', ["gini", "entropy"]), # default=gini
"max_depth": trial.suggest_categorical('max_depth', [None, 1, 2, 5, 10, 50]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# max_leaf_nodes
# min_impurity_decrease
# bootstrap
# oob_score
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.GradientBoostingClassifier:
return {
"loss": trial.suggest_categorical('loss', ["deviance", "exponential"]), # default=deviance
"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=0.1
"n_estimators": trial.suggest_int('n_estimators', 2, 200), # default=100
"subsample": trial.suggest_uniform('subsample', 0, 1), # default=1
"criterion": trial.suggest_categorical('criterion', ["friedman_mse", "mse"]), # default=friedman_mse
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
"max_depth": trial.suggest_int('max_depth', 1, 10), # default=3
# min_impurity_decrease
# init
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.RandomForestClassifier:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 150), # default=100
"criterion": trial.suggest_categorical('criterion', ["gini", "entropy"]), # default=gini
"max_depth": trial.suggest_categorical('max_depth', [None, 1, 2, 5, 10, 20]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# max_leaf_nodes
# min_impurity_decrease
# bootstrap
# oob_score
"random_state": random_seed
}, not_support_pattern
# Gaussian Processes
if model_cls == sklearn.gaussian_process.GaussianProcessClassifier:
return {"random_state": random_seed}, not_support_pattern
# GLM
if model_cls == sklearn.linear_model.LogisticRegression:
def not_support_pattern(params):
if params["penalty"] == "elasticnet":
if params["solver"] != "saga":
return True
if params["solver"] == "lbfgs":
if params["dual"] == True:
return True
if params["penalty"] == "l1":
return True
if params["solver"] == "saga":
if params["dual"] == True:
return True
if params["solver"] == "newton-cg":
if params["dual"] == True:
return True
if params["penalty"] not in ["l2", "none"]:
return True
if params["solver"] == "sag":
if params["penalty"] not in ["l2", "none"]:
return True
if params["dual"] == True:
return True
if params["solver"] == "liblinear":
if params["penalty"] == "none":
return True
return False
return {
"penalty": trial.suggest_categorical('penalty', ["l1", "l2", "elasticnet", "none"]), # default=l2
"dual": trial.suggest_categorical('dual', [False, True]), # default=False
# tol
"C": trial.suggest_categorical('C', [0.01, 0.1, 1, 2, 5]), # default=1.0
"solver": trial.suggest_categorical('solver', ["newton-cg", "lbfgs", "liblinear", "sag", "saga"]), # default=lbfgs
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.linear_model.RidgeClassifier:
return {
"alpha": trial.suggest_uniform('alpha', 0, 1), # default=1
"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
"normalize": trial.suggest_categorical('normalize', [False, True]), # default=False
"random_state": random_seed
}, not_support_pattern
#Navies Bayes
if model_cls == sklearn.naive_bayes.BernoulliNB:
return {
"alpha": trial.suggest_uniform('alpha', 0, 1), # default=1
}, not_support_pattern
if model_cls == sklearn.naive_bayes.GaussianNB:
return {}, not_support_pattern
# Nearest Neighbor
if model_cls == sklearn.neighbors.KNeighborsClassifier:
return {
"n_neighbors": trial.suggest_int('n_neighbors', 1, 100), # default=5
"weights": trial.suggest_categorical('weights', ["uniform", "distance"]), # default=uniform
"algorithm": trial.suggest_categorical('algorithm', ["auto", "ball_tree", "kd_tree", "brute"]), # default=auto
"leaf_size": trial.suggest_int('leaf_size', 1, 100), # default=30
"p": trial.suggest_categorical('p', [1, 2, 3, 4]), # default=2
# metric
# metric_params
}, not_support_pattern
# Trees
if model_cls == sklearn.tree.DecisionTreeClassifier:
return {
"criterion": trial.suggest_categorical('criterion', ["gini", "entropy"]), # default=gini
"splitter": trial.suggest_categorical('splitter', ["random", "best"]), # default=random
"max_depth": trial.suggest_categorical('max_depth', [None, 2, 10, 50, 100]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# min_impurity_decrease
# max_leaf_nodes
# ccp_alpha
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.tree.ExtraTreeClassifier:
return {
"criterion": trial.suggest_categorical('criterion', ["gini", "entropy"]), # default=gini
"splitter": trial.suggest_categorical('splitter', ["random", "best"]), # default=random
"max_depth": trial.suggest_categorical('max_depth', [None, 2, 10, 50, 100]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# min_impurity_decrease
# max_leaf_nodes
# ccp_alpha
"random_state": random_seed
}, not_support_pattern
# SVM
if model_cls == sklearn.svm.SVC:
return {
"C": trial.suggest_categorical('C', [0.01, 0.1, 0.5, 1.0, 2.0, 5.0]), # default=1.0
#"kernel": trial.suggest_categorical('kernel', ["linear", "poly", "rbf", "sigmoid", "precomputed"]), # default=rbf
"kernel": trial.suggest_categorical('kernel', ["linear", "poly", "rbf", "sigmoid"]), # default=rbf
# degree
"gamma": trial.suggest_categorical('gamma', ["auto", "scale"]), # default=scale
# coef0
"shrinking": trial.suggest_categorical('shrinking', [False, True]), # default=True
"probability": trial.suggest_categorical('probability', [False, True]), # default=False
# tol
# cache_size
# max_iter
"random_state": trial.suggest_categorical('random_state', [random_seed]),
}, not_support_pattern
if model_cls == sklearn.svm.NuSVC:
return {
"nu": trial.suggest_uniform('nu', 0, 1), # default=0.5
"kernel": trial.suggest_categorical('kernel', ["linear", "poly", "rbf", "sigmoid", "precomputed"]), # default=rbf
# degree
# gamma
# coef0
#"shrinking": trial.suggest_categorical('shrinking', [False, True]), # default=True
# tol
# cache_size
# max_iter
}, not_support_pattern
if model_cls == sklearn.svm.LinearSVC:
def not_support_pattern(params):
if params["penalty"] == "l1" and params["dual"] == True:
return True
if params["penalty"] == "l1" and params["loss"] == "hinge":
return True
if params["penalty"] == "l2" and params["loss"] == "hinge" and params["dual"] == False:
return True
return False
return {
"penalty": trial.suggest_categorical('penalty', ["l1", "l2"]), # default=l2
"loss": trial.suggest_categorical('loss', ["hinge", "squared_hinge"]), # default=squared_hinge
"tol": trial.suggest_categorical('tol', [1e-5, 1e-4, 1e-3]), # default=1e-4
"C": trial.suggest_categorical('C', np.logspace(-2, 1, 30)), # default=1.0
"dual": trial.suggest_categorical('dual', [False, True]), # default=True
#"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
#"intercept_scaling":
"max_iter": trial.suggest_categorical('max_iter', [100, 1000, 2000]), # default=1000
"random_state": random_seed,
}, not_support_pattern
# NN
if model_cls == sklearn.neural_network.MLPClassifier:
return {
"hidden_layer_sizes": (100,), # default=(100,)
#"activation": trial.suggest_categorical('activation', ["identity", "logistic", "tanh", "relu"]), # default=relu
#"solver": trial.suggest_categorical('solver', ["lbfgs", "sgd", "adam"]), # default=adam
#"alpha": trial.suggest_uniform('alpha', 0, 0.1), # default=0.0001
"batch_size": trial.suggest_categorical('batch_size', ["auto", 32, 64, 128, 256, 512]), # default=auto
"learning_rate": trial.suggest_categorical('learning_rate', ["constant", "invscaling", "adaptive"]), # default=constant
"learning_rate_init": trial.suggest_uniform('learning_rate_init', 0, 0.1), # default=0.001
#"power_t":
"max_iter": trial.suggest_categorical('max_iter', [100, 200, 500]), # default=200
"random_state": random_seed
}, not_support_pattern
#xgboost
if model_cls == xgb.XGBClassifier:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=
#"max_depth": trial.suggest_categorical('max_depth', [-1, 5, 10, 50, 100]), # default=
"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=
"eval_metric": "logloss",
"use_label_encoder": False,
"random_state": random_seed,
}, not_support_pattern
# light gbm
if model_cls == lgb.LGBMClassifier:
return {
"boosting_type": trial.suggest_categorical('boosting_type', ["gbdt", "dart", "goss"]), # default=gbdt
"num_leaves": trial.suggest_categorical('num_leaves', [2, 10, 31, 50, 100]), # default=31
"max_depth": trial.suggest_categorical('max_depth', [-1, 5, 10, 50, 100]), # default=-1
"learning_rate": trial.suggest_categorical('learning_rate', [0.001, 0.01, 0.1]), # default=0.1
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=1000
#"subsample_for_bin": trial.suggest_int('subsample_for_bin', 100, 1000000), # default=200000
#"objective": trial.suggest_categorical('objective', [None]), # default=None
# class_weight
#"min_split_gain": trial.suggest_uniform('min_split_gain', 0, 1), # default=0
#"min_child_weight": trial.suggest_uniform('min_child_weight', 0, 1), # default=1e-3
#"min_child_samples ": trial.suggest_int('min_child_samples ', 1, 100), # default=20
#"subsample": trial.suggest_uniform('subsample', 0, 1), # default=1
#"subsample_freq": trial.suggest_int('subsample_freq ', 0, 100), # default=0
#"colsample_bytree"
#"reg_alpha": trial.suggest_uniform('reg_alpha', 0, 1), # default=0
#"reg_lambda": trial.suggest_uniform('reg_lambda', 0, 1), # default=0
"random_state": random_seed
}, not_support_pattern
# catboost
if model_cls == catboost.CatBoostClassifier:
return {
"iterations": trial.suggest_int('iterations', 100, 2000), # default=1000
"depth": trial.suggest_int('depth', 4, 10), # default=6
"l2_leaf_reg": trial.suggest_int('l2_leaf_reg', 1, 10), # default=3
#"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=0.03
"random_strength": trial.suggest_int('random_strength', 1, 10), # default=1
"bagging_temperature": trial.suggest_categorical('bagging_temperature', [0, 0.5, 1, 10, 100]), # default=1
"has_time": trial.suggest_categorical('has_time', [False, True]), # default=False
"random_state": trial.suggest_categorical('random_state', [random_seed]),
"verbose": trial.suggest_categorical('verbose', [0])
}, not_support_pattern
#-----------------------------------------------------------
# Regressor
#-----------------------------------------------------------
#Ensemble Methods
if model_cls == sklearn.ensemble.AdaBoostRegressor:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=50
"learning_rate": trial.suggest_uniform('learning_rate', 0, 1), # default=1
"loss": trial.suggest_categorical('loss', ["linear", "square", "exponential"]), # default=linear
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.BaggingRegressor:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 500), # default=10
"max_samples": trial.suggest_uniform('max_samples', 0, 1), # default=1
"max_features": trial.suggest_uniform('max_features', 0, 1), # default=1
"bootstrap": trial.suggest_categorical('bootstrap', [False, True]), # default=True
"bootstrap_features": trial.suggest_categorical('bootstrap_features', [False, True]), # default=False
# oob_score
# warm_start
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.ExtraTreesRegressor:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 100), # default=100
"criterion": trial.suggest_categorical('criterion', ["mse", "mae"]), # default=mse
"max_depth": trial.suggest_categorical('max_depth', [None, 1, 2, 5, 10, 50]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# max_leaf_nodes
# min_impurity_decrease
# bootstrap
# oob_score
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.GradientBoostingRegressor:
return {
"loss": trial.suggest_categorical('loss', ["ls", "lad", "huber", "quantile"]), # default=ls
"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=0.1
"n_estimators": trial.suggest_int('n_estimators', 2, 200), # default=100
"subsample": trial.suggest_uniform('subsample', 0, 1), # default=1
"criterion": trial.suggest_categorical('criterion', ["friedman_mse", "mse"]), # default=friedman_mse
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
"max_depth": trial.suggest_int('max_depth', 1, 10), # default=3
# min_impurity_decrease
# init
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.ensemble.RandomForestRegressor:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 150), # default=100
"criterion": trial.suggest_categorical('criterion', ["mse", "mae"]), # default=mse
"max_depth": trial.suggest_categorical('max_depth', [None, 1, 2, 5, 10, 20]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# max_leaf_nodes
# min_impurity_decrease
# bootstrap
# oob_score
"random_state": random_seed
}, not_support_pattern
# GLM
if model_cls == sklearn.linear_model.Lasso:
return {
"alpha": trial.suggest_uniform('alpha', 0, 1), # default=1
"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
"normalize": trial.suggest_categorical('normalize', [False, True]), # default=False
"random_state": trial.suggest_categorical('random_state', [random_seed]), # default=None
}, not_support_pattern
if model_cls == sklearn.linear_model.Ridge:
return {
"alpha": trial.suggest_uniform('alpha', 0, 1), # default=1
"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
"normalize": trial.suggest_categorical('normalize', [False, True]), # default=False
"random_state": trial.suggest_categorical('random_state', [random_seed]), # default=None
}, not_support_pattern
if model_cls == sklearn.linear_model.ElasticNet:
return {
"alpha": trial.suggest_uniform('alpha', 0, 1), # default=1
"l1_ratio": trial.suggest_uniform('l1_ratio', 0, 1), # default=0.5
"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
"normalize": trial.suggest_categorical('normalize', [False, True]), # default=False
"random_state": trial.suggest_categorical('random_state', [random_seed]), # default=None
}, not_support_pattern
# Nearest Neighbor
if model_cls == sklearn.neighbors.KNeighborsRegressor:
return {
"n_neighbors": trial.suggest_int('n_neighbors', 1, 100), # default=5
"weights": trial.suggest_categorical('weights', ["uniform", "distance"]), # default=uniform
"algorithm": trial.suggest_categorical('algorithm', ["auto", "ball_tree", "kd_tree", "brute"]), # default=auto
"leaf_size": trial.suggest_int('leaf_size', 1, 100), # default=30
"p": trial.suggest_categorical('p', [1, 2, 3, 4]), # default=2
# metric
# metric_params
}, not_support_pattern
# Trees
if model_cls == sklearn.tree.DecisionTreeRegressor:
return {
"criterion": trial.suggest_categorical('criterion', ["mse", "friedman_mse", "mae"]), # default=mse
"splitter": trial.suggest_categorical('splitter', ["random", "best"]), # default=random
"max_depth": trial.suggest_categorical('max_depth', [None, 2, 10, 50, 100]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# min_impurity_decrease
# max_leaf_nodes
# ccp_alpha
"random_state": random_seed
}, not_support_pattern
if model_cls == sklearn.tree.ExtraTreeRegressor:
return {
"criterion": trial.suggest_categorical('criterion', ["mse", "friedman_mse", "mae"]), # default=mse
"splitter": trial.suggest_categorical('splitter', ["random", "best"]), # default=random
"max_depth": trial.suggest_categorical('max_depth', [None, 2, 10, 50, 100]), # default=None
# min_samples_split
# min_samples_leaf
# min_weight_fraction_leaf
# max_features
# min_impurity_decrease
# max_leaf_nodes
# ccp_alpha
"random_state": random_seed
}, not_support_pattern
# SVM
if model_cls == sklearn.svm.SVR:
return {
"kernel": trial.suggest_categorical('kernel', ["linear", "poly", "rbf"]), # default=rbf
# degree
# gamma
# coef0
# tol
"C": trial.suggest_categorical('C', [0.01, 0.1, 1, 2, 10]), # default=1.0
"epsilon": trial.suggest_uniform('epsilon', 0, 1), # default=0.1
"shrinking": trial.suggest_categorical('shrinking', [False, True]), # default=True
# cache_size
# max_iter
}, not_support_pattern
if model_cls == sklearn.svm.NuSVR:
return {
"nu": trial.suggest_uniform('nu', 0, 1), # default=0.5
"C": trial.suggest_categorical('C', [0.01, 0.1, 1, 2, 10]), # default=1.0
"kernel": trial.suggest_categorical('kernel', ["linear", "poly", "rbf"]), # default=rbf
# degree
# gamma
# coef0
"shrinking": trial.suggest_categorical('shrinking', [False, True]), # default=True
# tol
# cache_size
# max_iter
}, not_support_pattern
if model_cls == sklearn.svm.LinearSVR:
return {
#"epsilon": trial.suggest_uniform('epsilon', 0, 1), # default=0
#"tol": trial.suggest_uniform('tol', 0, 1), # default=1e-4
"C": trial.suggest_uniform('C', 0, 1), # default=1.0
"loss": trial.suggest_categorical('loss', ["epsilon_insensitive", "squared_epsilon_insensitive"]), # default=epsilon_insensitive
"fit_intercept": trial.suggest_categorical('fit_intercept', [False, True]), # default=True
#"intercept_scaling":
#"dual": trial.suggest_categorical('dual', [False, True]), # default=True
"max_iter": trial.suggest_categorical('max_iter', [100, 1000, 10000]), # default=1000
}, not_support_pattern
# NN
if model_cls == sklearn.neural_network.MLPRegressor:
return {
"hidden_layer_sizes": (100,), # default=(100,)
#"activation": trial.suggest_categorical('activation', ["identity", "logistic", "tanh", "relu"]), # default=relu
#"solver": trial.suggest_categorical('solver', ["lbfgs", "sgd", "adam"]), # default=adam
#"alpha": trial.suggest_uniform('alpha', 0, 0.1), # default=0.0001
"batch_size": trial.suggest_categorical('batch_size', ["auto", 32, 64, 128, 256, 512]), # default=auto
"learning_rate": trial.suggest_categorical('learning_rate', ["constant", "invscaling", "adaptive"]), # default=constant
"learning_rate_init": trial.suggest_uniform('learning_rate_init', 0, 0.1), # default=0.001
#"power_t":
"max_iter": trial.suggest_categorical('max_iter', [100, 200, 500]), # default=200
"random_state": random_seed
}, not_support_pattern
#xgboost
if model_cls == xgb.XGBRegressor:
return {
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=
#"max_depth": trial.suggest_categorical('max_depth', [-1, 5, 10, 50, 100]), # default=
"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=
"objective": 'reg:squarederror',
"eval_metric": "logloss",
"use_label_encoder": False,
"random_state": random_seed,
}, not_support_pattern
# light bgm
if model_cls == lgb.LGBMRegressor:
return {
"boosting_type": trial.suggest_categorical('boosting_type', ["gbdt", "dart", "goss"]), # default=gbdt
"num_leaves": trial.suggest_categorical('num_leaves', [2, 10, 31, 50, 100]), # default=31
"max_depth": trial.suggest_categorical('max_depth', [-1, 5, 10, 50, 100]), # default=-1
"learning_rate": trial.suggest_categorical('learning_rate', [0.001, 0.01, 0.1]), # default=0.1
"n_estimators": trial.suggest_int('n_estimators', 2, 1000), # default=1000
#"subsample_for_bin": trial.suggest_int('subsample_for_bin', 100, 1000000), # default=200000
#"objective": trial.suggest_categorical('objective', [None]), # default=None
# class_weight
#"min_split_gain": trial.suggest_uniform('min_split_gain', 0, 1), # default=0
#"min_child_weight": trial.suggest_uniform('min_child_weight', 0, 1), # default=1e-3
#"min_child_samples ": trial.suggest_int('min_child_samples ', 1, 100), # default=20
#"subsample": trial.suggest_uniform('subsample', 0, 1), # default=1
#"subsample_freq": trial.suggest_int('subsample_freq ', 0, 100), # default=0
#"colsample_bytree"
#"reg_alpha": trial.suggest_uniform('reg_alpha', 0, 1), # default=0
#"reg_lambda": trial.suggest_uniform('reg_lambda', 0, 1), # default=0
"random_state": random_seed
}, not_support_pattern
# catboost
if model_cls == catboost.CatBoostRegressor:
return {
"iterations": trial.suggest_int('iterations', 100, 2000), # default=1000
"depth": trial.suggest_int('depth', 4, 10), # default=6
"l2_leaf_reg": trial.suggest_int('l2_leaf_reg', 1, 10), # default=3
#"learning_rate": trial.suggest_uniform('learning_rate', 0, 0.1), # default=0.03
"random_strength": trial.suggest_int('random_strength', 1, 10), # default=1
"bagging_temperature": trial.suggest_categorical('bagging_temperature', [0, 0.5, 1, 10, 100]), # default=1
"has_time": trial.suggest_categorical('has_time', [False, True]), # default=False
"random_state": trial.suggest_categorical('random_state', [random_seed]),
"verbose": trial.suggest_categorical('verbose', [0])
}, not_support_pattern
パラメータ調整コード例
import optuna
# optuna のINFO以下を非表示
optuna.logging.set_verbosity(optuna.logging.WARNING)
# 警告非表示
import warnings
warnings.simplefilter('ignore')
from tqdm import tqdm
import time
# 結果格納用
df_compare = pd.DataFrame(columns=['name', 'metric1', "metric2", 'params', 'time'])
# ---------------------------
# optunaの定義
# ---------------------------
def objective_degree(model_cls, df):
def objective(trial):
params, not_support_pattern = create_optuna_params(trial, model_cls, 1234)
if not_support_pattern is not None:
# サポートされていない組み合わせは最低評価
if not_support_pattern(params):
return 0
try:
#--- モデルを評価する
metric = validation_classifier(
df, select_columns, "Survived", model_cls, params)
except Exception as e:
print("---------------")
print("error")
print(model_cls)
print(params)
print(e)
print("---------------")
metric = 0
return metric
return objective
# ---------------------------
# モデル毎にパラメータをサーチする
# ---------------------------
for model_ in tqdm(get_models_classifier(1234)):
model_cls = model_[0]
model_params = model_[1]
name = model_cls.__name__
# デフォルトパラメータでの結果を取得しておく
metric1 = validation_classifier(
df, select_columns, "Survived", model_cls, model_params)
# 調整開始
t0 = time.time()
study = optuna.create_study(direction="maximize")
study.optimize(objective_degree(model_cls, df),
timeout=60*1, n_trials=1000)
# optunaの結果を取得
best_value = study.best_value
best_params = study.best_params
trial_count = len(study.trials)
t1 = time.time() - t0
# 結果を保存
df_compare = df_compare.append({
"name": name,
"metric1": metric1,
"metric2": best_value,
"params": best_params,
"count": trial_count,
"time": t1,
}, ignore_index=True)
# 結果を表示
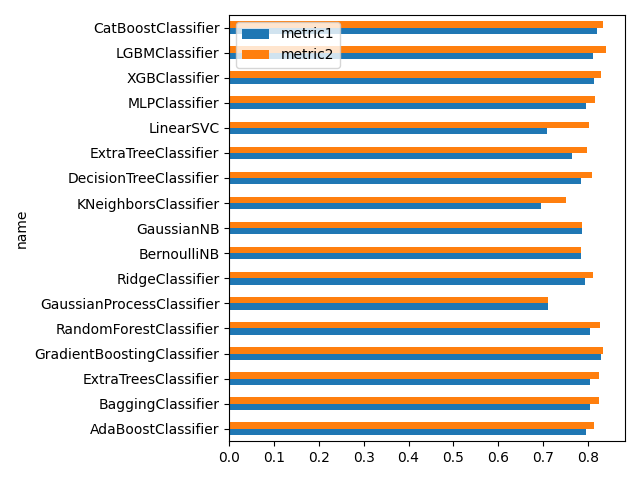
print(df_compare)
df_compare.set_index('name', inplace=True)
df_compare.plot.barh(y=["metric1", "metric2"])
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
name metric1 metric2 time count
0 AdaBoostClassifier 0.795735 0.813692 60.745781 14.0
1 BaggingClassifier 0.804714 0.824916 60.128095 24.0
2 ExtraTreesClassifier 0.804714 0.823793 60.199412 227.0
3 GradientBoostingClassifier 0.829405 0.832772 60.605364 48.0
4 RandomForestClassifier 0.804714 0.826038 60.197980 67.0
5 GaussianProcessClassifier 0.710438 0.710438 60.484813 77.0
6 RidgeClassifier 0.792368 0.810325 60.332088 801.0
7 BernoulliNB 0.784512 0.784512 50.800641 1000.0
8 GaussianNB 0.786756 0.786756 38.703743 1000.0
9 KNeighborsClassifier 0.694725 0.751964 60.365270 638.0
10 DecisionTreeClassifier 0.784512 0.808081 60.623820 968.0
11 ExtraTreeClassifier 0.764310 0.797980 60.452333 987.0
12 LinearSVC 0.708193 0.801347 60.207922 134.0
13 MLPClassifier 0.794613 0.815937 60.385311 50.0
14 XGBClassifier 0.812570 0.829405 60.495781 41.0
15 LGBMClassifier 0.810325 0.840629 60.635715 43.0
16 CatBoostClassifier 0.819304 0.832772 66.098995 5.0
metric1がデフォルト値、metric2が調整後の数値です。
ハイパーパラメータ調整の効果はあるようです。
ただ、時間がかなりかかりますね…
8.その他のテクニック
目的変数の対数変換(回帰)
目的変数の対数変換です。
参照:どのようなときに目的変数Yではなくlog(Y)にしたほうがよいのか?~対数変換するメリットとデメリット~
参照のメリットを引用すると、
メリット: Y の値が小さいサンプルの誤差が小さくなる
デメリット: Y の値が大きいサンプルの誤差が大きくなる
です。
- 対数変換
df["Survived"] = np.log(df["Survived"])
- 元に戻す
model = 対数変換した目的変数でモデルを学習
# 予測する
x = df[df["Survived"].isnull()][select_columns]
y_pred = model.predict(x)
# 予測結果を元に戻す
y_pred = np.exp(y_pred)
例のSurvivedは分類問題なので実施するメリットはありませんけど…
9.AutoML(PyCaret)の適用
PyCaret を使う例です。
表示はJupyter Notebookで使う場合の出力に最適化されているようです。
1.データ読み込み
学習に使うデータは以下です。
pcc_df = df[df["Survived"].notnull()][[
"Pclass",
"Sex",
"Age",
"SibSp",
"Parch",
"Family",
"Fare",
"Embarked",
"Survived", # 学習データなので目的変数もいれます
]]
# データを渡す
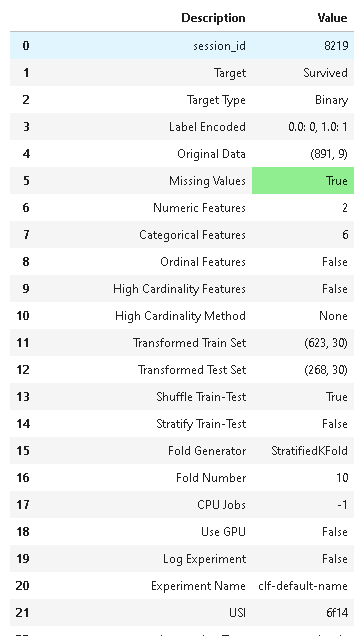
import pycaret.classification as pcc
r = pcc.setup(pcc_df, target="Survived", silent=True)
結果は59項目ありました。
重要な項目は色がつくようです?
2.モデル選択
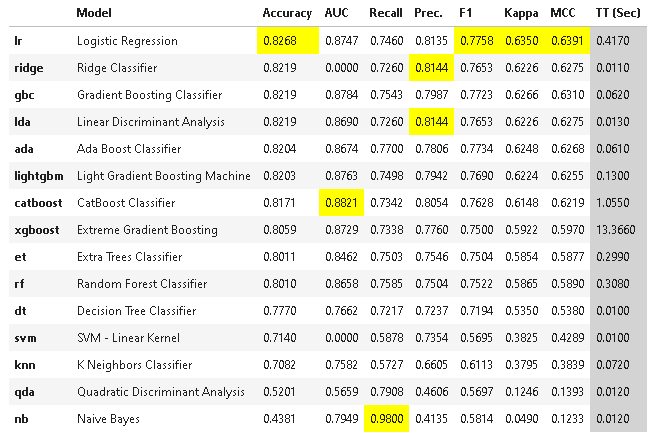
- 複数のモデルを比較
数分かかります。
best_model = pcc.compare_models()
print(best_model.__class__.__name__) # LogisticRegression
- 任意のモデル
任意のモデルを選びたい場合はこちらから
model = pcc.create_model("lightgbm")
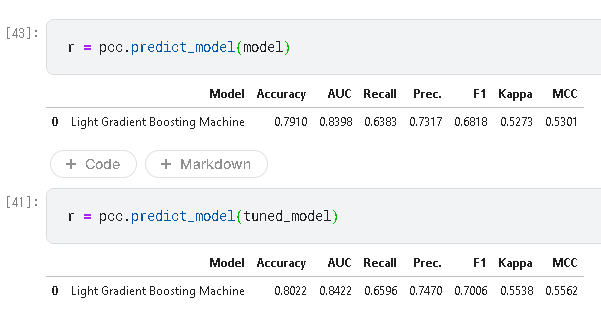
3.ハイパーパラメータの調整
tuned_model = pcc.tune_model(model)
print(tuned_model.get_params)
<bound method LGBMModel.get_params of LGBMClassifier(bagging_fraction=0.7, bagging_freq=3, boosting_type='gbdt',
class_weight=None, colsample_bytree=1.0, feature_fraction=0.8,
importance_type='split', learning_rate=0.1, max_depth=-1,
min_child_samples=36, min_child_weight=0.001, min_split_gain=0.4,
n_estimators=230, n_jobs=-1, num_leaves=50, objective=None,
random_state=8219, reg_alpha=0.1, reg_lambda=3, silent=True,
subsample=1.0, subsample_for_bin=200000, subsample_freq=0)>
調整前後の評価値です。
predict_model はdataを指定しないとホールドアウトで実行した結果を返します。
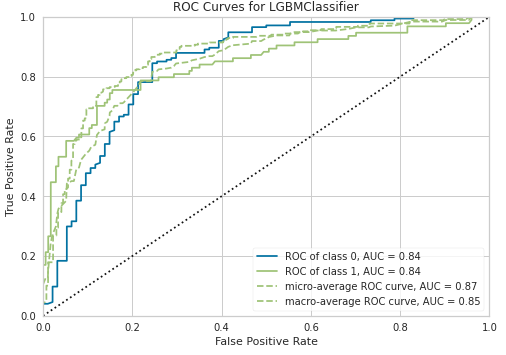
4.モデルの可視化
pcc.plot_model(tuned_model)
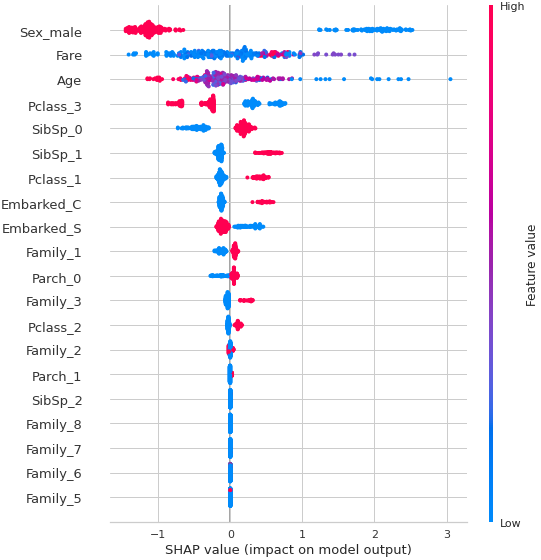
5.モデル解釈の可視化
pcc.interpret_model(tuned_model)
6.予測
"Label"と"Score"カラムが追加され、Labelは予測結果、分類なら各クラスの予測確率がScoreに入っています。
x_pred = df[df["Survived"].isnull()]
y_pred = pcc.predict_model(tuned_model, data=x_pred)
print(y_pred["Label"])
891 0.0
892 0.0
893 0.0
894 0.0
895 1.0
...
1304 0.0
1305 1.0
1306 0.0
1307 0.0
1308 0.0
Name: Label, Length: 418, dtype: object