こんにちは。
バイオインフォマティクスネタです。TCGAの公開している突然変異のデータにNMF(非負値行列因子分解)をかけたら、変異のシグネチャーがとれるかな?と単純に考えてやってみたところ、割とうまくいきましたので記事にします。
気づいたことはコメントや編集リクエストで指摘して頂けるとうれしいです。
🐣 〜基礎知識編〜
🌽 変異シグネチャーってなに?
がん細胞の遺伝子変異にはパターンがある
がんは遺伝子の病気であると言われています。がん細胞には多数の遺伝子変異があり、増殖、免疫の回避、独自の代謝など、がん細胞としての性質を獲得しているとされます。発がんにいたるまでに細胞はさまざまな遺伝子変異を蓄積します。遺伝子変異の原因としてタバコ・紫外線などの発がん物質が有名ですが、DNAの損傷を修復する機構の破綻も影響しているとされます。
正常細胞からがん細胞ができるまでには長い時間をかけて環境に適した細胞が選択されていく必要があります。
蓄積された変異の頻度を調べて、タバコにはタバコ、紫外線には紫外線の特徴的なパターンを探すのがシグネチャーの考え方です。
遺伝子変異のパターンを分解する
がん細胞の突然変異は複数の原因が組み合わさって発生しています。これを分解するためにNMF(非負値行列因子分解)という手法を使います。
Single Base Substitution (SBS) Signatures
変異シグネチャーのなかで、Single Base Substitution (SBS) Signaturesが最も有名です。その他にもシグネチャーはいろいろあるようですが、私は詳しくありません。
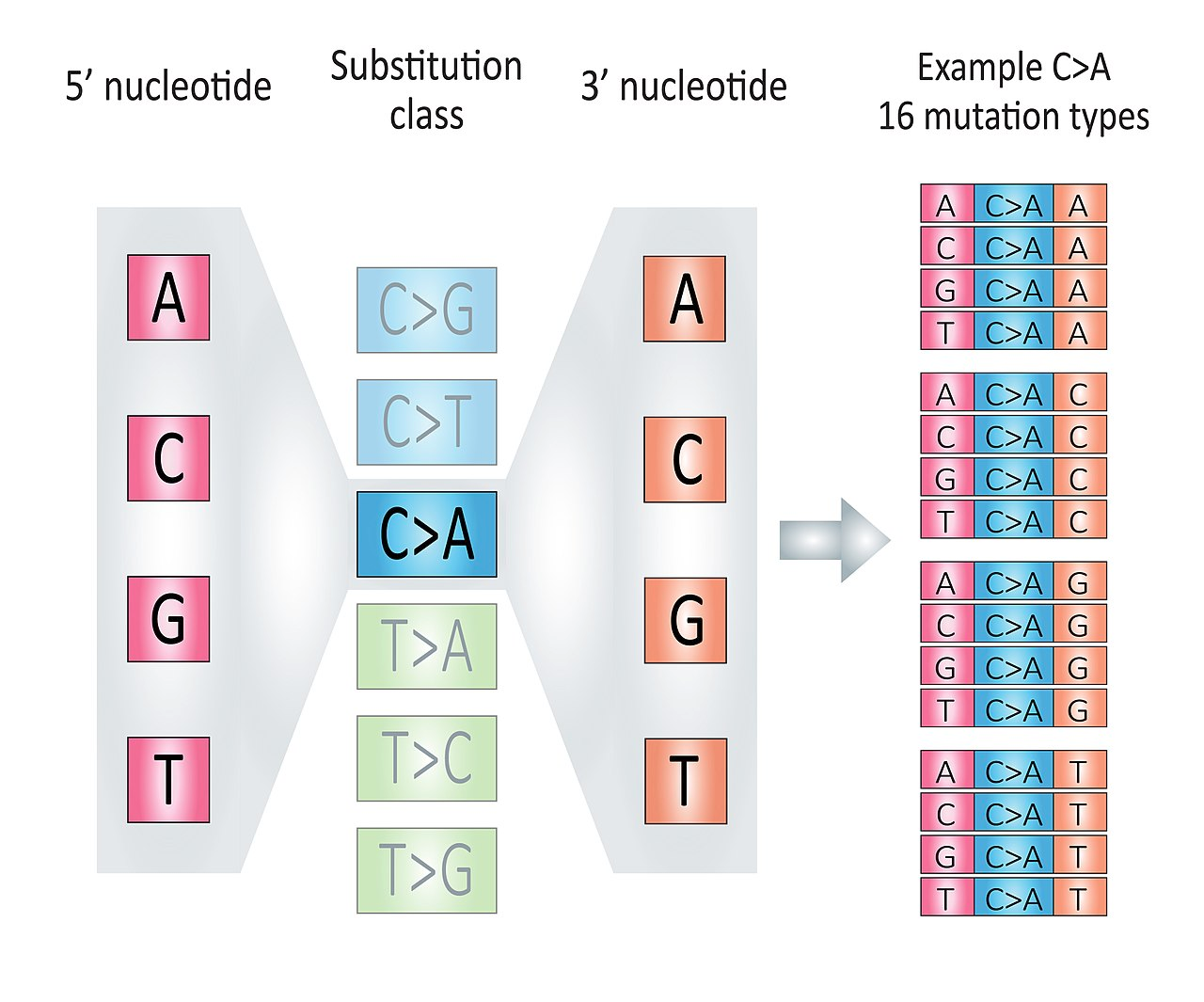
このシグネチャーは点変異と、その左右の塩基をカウントします。
- 塩基の変異のパターン
- 5'末端側の塩基
- 3'末端側の塩基
の場合の数は全部で48通りあります。この48通りについて頻度をカウントして行列を作ってシグネチャーとして定義します。
(Wikipedia https://en.wikipedia.org/wiki/Mutational_signatures より)
これだけではよくわからないと思うので、次に具体例を示します。
🌌 COSMICの変異シグネチャーをながめる
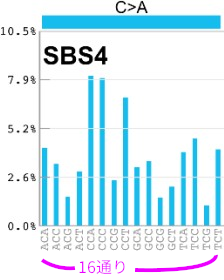
https://cancer.sanger.ac.uk/cosmic/signatures/SBS/SBS4.tt
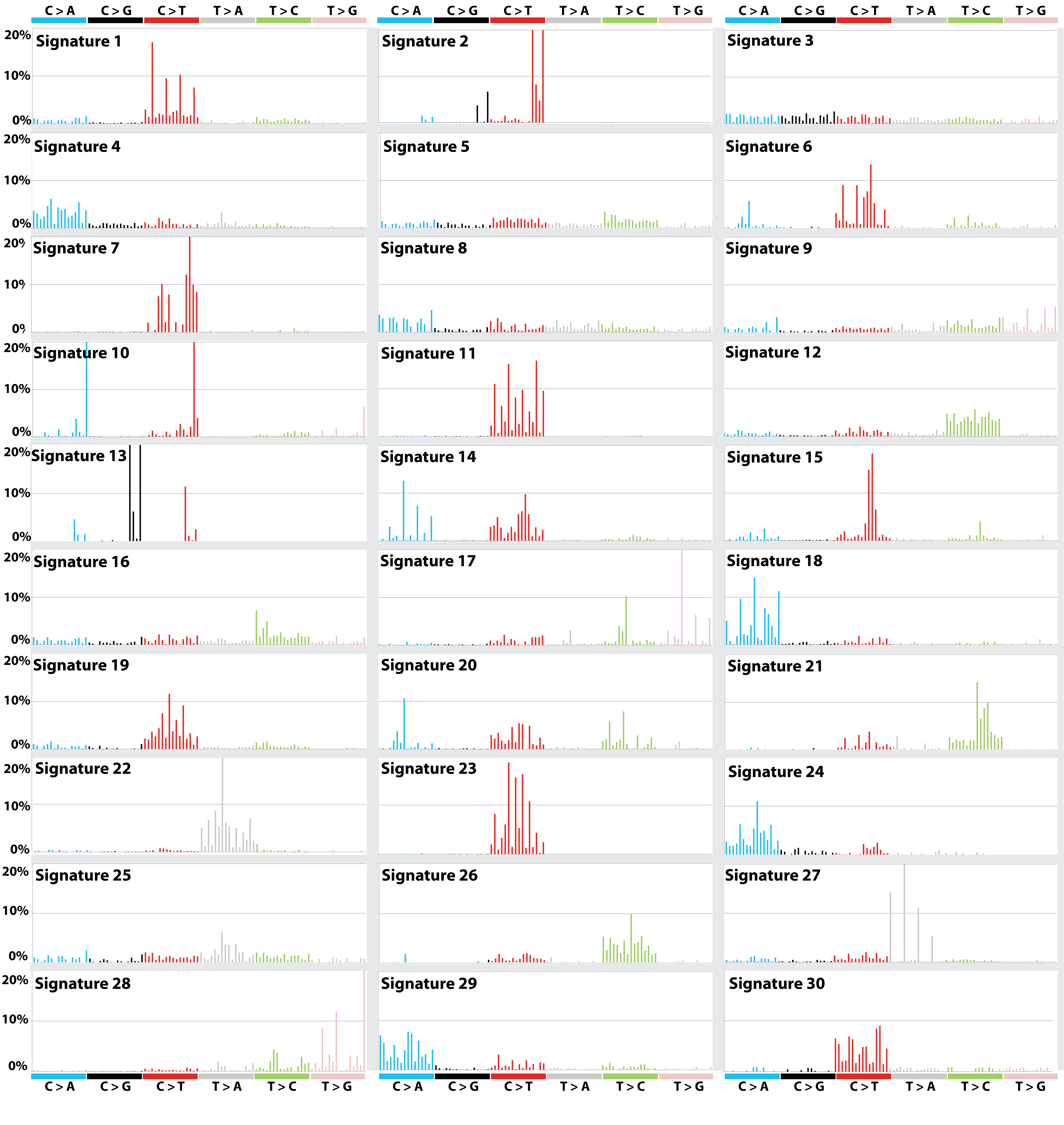
下の図に示したのがSignature4です。4番はタバコの有害物質の一つ、ベンゾピレンの変異シグネチャーです。
それぞれのColumn(列)のバーが、変異の頻度を示しています。左側の水色のバーはC→Aへの変異の頻度を示しています。黒はC→Gの変異、赤はC→Tの変異、灰色はT→Aの変異、黄緑色はT→Cの変異、ピンクがT→Gの変異を示しています。点変異パターンが6種類しかないことに気が付かれましたでしょうか。たとえばG→Tへの変異というパターンがありません。なぜかというと、それは相補的な塩基から見てG→T変異は、C→A変異と同じものであるからです。よってG→Tは、C→Aと同一のものとしてカウントします。
この2つは同じこと
A G>T C →
| | | |
T C>A G ←
5'末端側が、A,C,G,Tの4通り、3'末端側もA,C,G,Tの4通りで、合計16通りの変異パターンがあります。CCAがCAAになる頻度が高く、TCGがTAGになる頻度はあまり高くないようですね。
Signatureの全体像を把握するためにCosmicに掲載されている30個のシグネチャー(Version2)を示します。
(最新版Version3はホームページを参照してください。)
🍅 〜目標〜
さて、今回はTCGAの突然変異データを使って塩基置換の行列式をつくり、Rumaleという機械学習ライブラリを使ってNMF(非負値行列因子分解)を実行します。これでCOSMICのシグネチャーが一部だけでも再現できたら成功ということにします。
突然変異のデータはcBioPortalで一般公開されているExonのものを使用します。TCGAの変異のデータはcBioPortalのAPI叩くだけで取得できます。(ただしWhole Genomeではないので精度は低くなりバイアスも入ると思います)
🍝 〜方法などについて〜
個別の方法について、過去のQiitaのエントリーへのリンクなど貼っておきます。
RubyでTCGAのデータベース cBioPortal から情報を取る方法
daru-apiclientという超薄い自作ツールでBioPortalからデータを取得しています。
httpartyでデータを取得しDaruにデータを渡しています。より詳しい情報は下記のページをご覧ください。
Rubyでがんのゲノム情報を機械学習してみた
https://github.com/kojix2/daru-apiclient
NMFについて
数学まったくわかりません。アルゴリズムの内容はわからないです。
他の方の書かれたわかりすい記事やネットの記事を参照して雰囲気でやっています。
非負値行列因子分解(NMF)をふわっと理解する
RubyでNMFを行う方法
RubyでNMFを実行する方法についてはこちらの記事を参照ください。
Rubyで顔のデータセットをNMF(非負値行列因子分解)してみた
RubyでT-SNEを行う方法
【Ruby】t-SNEでMNISTを次元圧縮して可視化してみた
Rumaleを実行するときはnumo-linalgをインストールしよう
RumaleでNMFやt-SNEを実行する場合は、numo-linalgの導入をオススメします。numo-linalgをインストールしなくてもRumaleは問題なく実行できるのですが、圧倒的な計算時間の差が発生します。特にコア数が多いCPUではその差が顕著です。numo-linalgは意外とインストールが難しいライブラリです。ソースコードからOpenBLASをインストールして、gem install -- --with-blas-dir オプションを指定するのが正攻法だと思います。しかし環境によってはOpenBLASをパッケージから入れて、単にgem install するだけでもインストールできるかも知れません。
numo-linalgをmacで動かしてみた
RubyでPlotlyを使う方法
超マイナーな方法になりますが、iruby-plotlyを使用しています。
控えめに言ってもめちゃくちゃ便利です。流行ってなくても便利なものは便利。
Rubyでバイオリンプロットを作る【Plotly】
https://github.com/zach-capalbo/iruby-plotly
ゲノムの任意の位置の塩基配列を取得する方法
@percipere さまの下記の記事をもとにsamtoolsを使用しました
ゲノムの任意の位置の塩基配列を取得したい
以下、「データ準備編」と「クラスタリング&可視化編」の2つのJupyter Notebookに分けて実行しています。
🏭 〜データ準備編〜
🐠 目次
準備
- 🔖 セーブポイント用関数を準備する
- 📖 ライブラリを読み込む
データ取得編
- 🌏 TCGAに接続する準備をする
- 📡 TCGAのスタディのIDを取得する
- 📡 Mutationのデータを取得する
前処理編
- 🔧 Mutationのデータを一つのDaru🍺ファイルにまとめる
- 💾 Mutationのデータを保存する
- 🔧 SNP かつ Chromosome 特定できるMutationを抽出する
- 💾 SNPのデータを保存する
Samtools編
- ✅ samtools の動作確認をする
- ✅ 両隣の塩基を確認する前に、ncbiBuildのバージョンが37であることを確認しておく
- 🔧 左右の塩基をsamtoolsで特定する
- 📚 mutations_snp_trinucleotide のデータ構造の確認
- 💾 トリヌクレオチドのデータを保存する
変異頻度カタログ作成編
- 🔧 シグネチャーの変異パターンを生成する
- 🔧 逆相補的配列を求める関数
- 🔧 シグネチャーの変異パターン(反対側)を生成する
- 🔧 インデックスラベルを生成する
- 🔧 変異の頻度をカウントしてカタログを作る
- 🔧 カタログをNArrayに変換する
- 💾 catalogのデータを保存する
NMF実行編
- 📚 RumaleのNMFの使い方を確認する
- 🔧 RumaleでNMFを実行して 💾 モデルを保存する
0. 🔖 セーブポイント用関数を準備する
## 今回は Signature ディレクトリにオブジェクトを保存していく
require 'fileutils'
FileUtils.mkdir_p("Signature")
def sync(str, obj)
begin
if obj.nil?
Marshal.load(File.binread("Signature/#{str}.dat"))
else
File.binwrite("Signature/#{str}.dat", Marshal.dump(obj))
obj
end
rescue
Marshal.load(File.binread("Signature/#{str}.dat"))
end
end
1. 📖 ライブラリを読み込む
require 'daru/apiclient' # TCGAのデータを取得します
require 'numo/linalg' # OpenBLAS
require 'rumale' # 機械学習
require 'parallel' # 並列計算
require 'iruby-plotly' # 可視化
require 'awesome_print' # 出力をきれいに
require 'rainbow' # 出力をきれいに
# Paralell で使用するコア数の設定
# Ryzen7のマシンで実行したので15で設定しました
PCORE = 15
15
2. 🌏 TCGAに接続する準備をする
c = Daru::APIClient.new "https://www.cbioportal.org/api"
<Daru::APIClient:0x00005588fd84ff40 @c=#<#:0x00005588fd84ee38>>
3. 📡 TCGAのスタディのIDを取得する
study_ids = c.get("/molecular-profiles").filter_rows{|r| r["molecularAlterationType"] == "MUTATION_EXTENDED"}
.filter_rows{|r| r["molecularProfileId"].include? "tcga"}["molecularProfileId"]
.to_a.find_all{|i| i.include? "_tcga_mutations"}
.map{|i| i.gsub("_mutations","")}
["acc_tcga", "blca_tcga", "brca_tcga", "cesc_tcga", "chol_tcga", "coadread_tcga", "dlbc_tcga", "esca_tcga", "gbm_tcga", "hnsc_tcga", "kich_tcga", "kirc_tcga", "kirp_tcga", "laml_tcga", "lgg_tcga", "lihc_tcga", "luad_tcga", "lusc_tcga", "meso_tcga", "ov_tcga", "paad_tcga", "pcpg_tcga", "prad_tcga", "sarc_tcga", "skcm_tcga", "stad_tcga", "tgct_tcga", "thca_tcga", "thym_tcga", "ucec_tcga", "ucs_tcga", "uvm_tcga"]
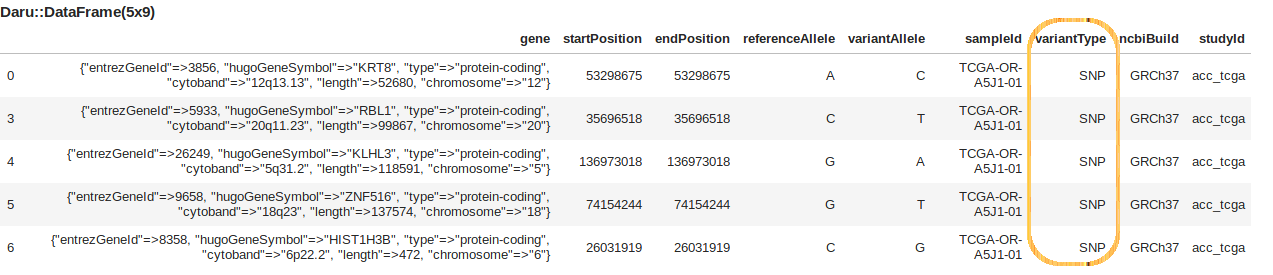
4. 📡 Mutationのデータを取得する
ここでは、TCGAのスタディごとにMutationのデータを取得します。
mutations = study_ids.map.with_index do |id, index|
puts [id, index]
df = c.get("/molecular-profiles/#{id}_mutations/mutations", query: {sampleListId: "#{id}_sequenced", projection: "DETAILED"})
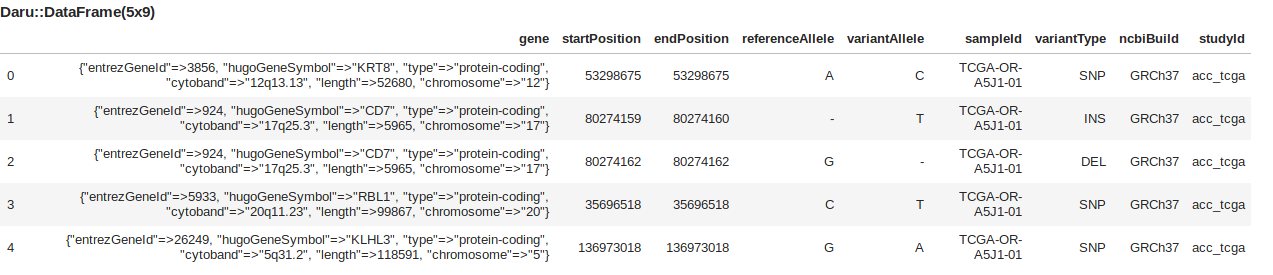
df2 = df["gene","startPosition","endPosition","referenceAllele","variantAllele","sampleId", "variantType", "ncbiBuild", "studyId"]
end
0
5. 🔧 Mutationのデータを一つのDaru🍺ファイルにまとめる
Daruは速度や信頼性の面で課題があるのですが、処理の流れのわかりやすさを考えてDaru::DataFrameにまとめました。
mutations2 = mutations.inject(&:concat)
# Daruの挙動を一応確認しておく
if mutations.map(&:size).sum == mutations2.size
puts "OK"
mutations = mutations2
mutations2 = nil # ほそぼそとしたメモリ対策
else
raise
end
# 先頭のデータを確認する
mutations.head(5)
# 総数
mutations.size
1118839
6. 💾 Mutationのデータを保存する
mutations = sync("mutations", mutations)
7. 🔧 SNP かつ Chromosome 特定できるMutationを抽出する
# SNPのみを選ぶ
mutations_snp = mutations.where(mutations.variantType.eq "SNP")
# Chromosome 特定できるMutationのみを選ぶ
# (keep_row_if メソッドは遅いのでfilter_rowsを使用)
mutations_snp = mutations_snp.filter_rows{|r| r["gene"]["chromosome"] != nil}
# 先頭のデータを確認する
mutations_snp.head(5)
8. 💾 SNPのデータを保存する
mutations_snp = sync("mutations_snp", mutations_snp)
💻 mutations変数をクリア
mutations = nil
🚀 Samtoolを使って、変異の両隣の塩基を特定する
9. ✅ samtools の動作確認をする
- @percipere さまの記事をもとにsamtoolsをセットアップしておく
- ゲノムの任意の位置の塩基配列を取得したい
puts `samtools faidx Homo_sapiens.GRCh37.dna.primary_assembly.fa.gz 7:150696110-150696118`
7:150696110-150696118
ATCCCCCAG
10. ✅ 両隣の塩基を確認する前に、ncbiBuildのバージョンが37であることを確認しておく
- GRCh37だけでなく、単なる37というのが含まれているが許容範囲でしょうかね
mutations_snp["ncbiBuild"].value_counts
11. 🔧 左右の塩基をsamtoolsで特定する
# IRubyでは Parallelで並列計算の最中に出力を行うとエラーが発生するので logger を準備する
require 'logger'
File.open("Signature/samtools.log",'w'){|file| file = nil}
logger = Logger.new("Signature/samtools.log")
start_time = Time.now
sample_ids = mutations_snp["sampleId"].to_a.uniq
mutations_snp_trinucleotide = Parallel.map(sample_ids, in_processes: PCORE, progress: "Samtools") do |sample_id|
# ここの効率よくないかも
mutations_per_sample_df = mutations_snp.where(mutations_snp["sampleId"].eq sample_id)
# Parallelでマルチコアを生かした計算をするため、Rubyの配列に変換する. 最後の to_h はなくても動く
# その場合Hashではなく、Daru::Vectorになる
mutations_per_sample = mutations_per_sample_df.each_row.to_a.map(&:to_h)
mutations_per_sample.map do |mutation|
chromosome = mutation["gene"]["chromosome"]
start_position = mutation["startPosition"]
end_position = mutation["endPosition"]
raise "start_position != end_position" if start_position != end_position
reference_allele = mutation["referenceAllele"]
variant_allele = mutation["variantAllele"]
trinucleotide = `samtools faidx Homo_sapiens.GRCh37.dna.primary_assembly.fa.gz \\
#{chromosome}:#{start_position-1}-#{end_position+1}`
.split("\n")[1]
# GRChの検索結果がTCGAの結果と一致していない場合ログを残す
# IRubyでは Parallelで並列計算の最中に出力を行うとエラーが発生するので logger を利用
unless trinucleotide.nil?
if trinucleotide[1] != reference_allele
logger.info "#{mutation["studyId"]} tri[1]:#{trinucleotide[1]} != ra:#{reference_allele}"
end
end
[trinucleotide, reference_allele, variant_allele]
end
end
puts time: (Time.now - start_time)
Samtools: |====================================================================|
{:time=>1287.954323263}
12. 📚 mutations_snp_trinucleotide のデータ構造の確認
puts mutations_snp_trinucleotide.class
puts "%d samples" % mutations_snp_trinucleotide.size
puts mutations_snp_trinucleotide[0]
13. 💾 トリヌクレオチドのデータを保存する
mutations_snp_trinucleotide = sync("mutations_snp_trinucleotide", mutations_snp_trinucleotide)
14. 🔧 シグネチャーの変異パターンを生成する
substitution_class = [['C' , 'A'],
['C' , 'G'],
['C' , 'T'],
['T' , 'A'],
['T' , 'C'],
['T' , 'G']]
ACGT = %w(A C G T)
mutation_types = []
substitution_class.each do |u,v|
ACGT.each do |j|
ACGT.each do |n|
mutation_types << ["" << j << u << n, v]
end
end
end
mutation_types = mutation_types.flatten(0)
15. 🔧 逆相補的配列を求める関数
def reverse_complement(str)
return nil if str == nil
complement = {"A" => "T", "T" => "A", "C" => "G", "G" => "C"}
str.reverse.chars.map do |s|
complement[s]
end.join
end
## 動作確認
reverse_complement("AAACTG")
"CAGTTT"
16. 🔧 シグネチャーの変異パターン(反対側)を生成する
mutation_types_reverse = mutation_types.map do |pattern|
pattern.map(&method(:reverse_complement))
end
17. 🔧 インデックスラベルを生成する
これは📊グラフの軸ラベルなどに使用するものです
signature_index = mutation_types.map{|i| i.join " > "}
File.binwrite("Signature/signature_index.dat", Marshal.dump(signature_index))
18. 🔧 変異の頻度をカウントしてカタログを作る
- カウントできずにエラーになったものが赤い文字で表示されます
mutations_snp_trinucleotide.sum(&:size)
catalog = mutations_snp_trinucleotide.map do |mutations_per_sample|
mutation_count = Array.new(96, 0)
mutations_per_sample.each do |mutation|
m = mutation.values_at(0,2)
index = mutation_types.index(m) || mutation_types_reverse.index(m)
if index.nil?
# 処理に失敗したものを表示する
print m.inspect.red
next
end
mutation_count[index] += 1
end
mutation_count
end
0
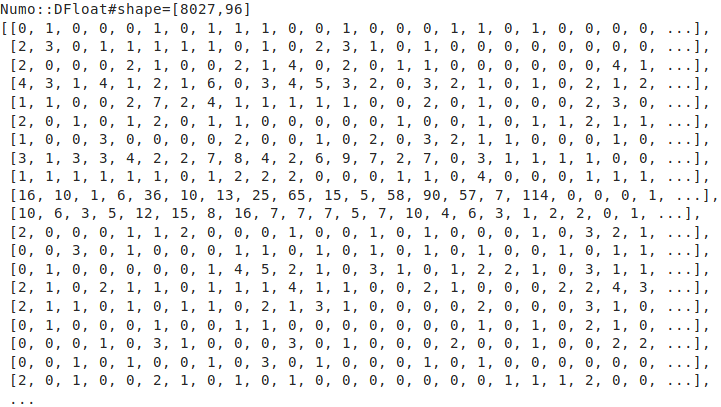
19. 🔧 NArrayに変換する
catalog = Numo::DFloat[*catalog]
puts catalog.inspect
20. 💾 catalogのデータを保存する
catalog = sync("catalog", catalog)
21. 📚 RumaleのNMFの使い方を確認する
ri Rumale::Decomposition::NMF.new
22. 🔧 RumaleでNMFを実行して 💾 モデルを保存する
- NMFでは局所解しか求まらないので、NMF500回試行してみることにします。
- コンポーネントの数は30とします。
- modelを保存します
- ここでは他のツールや他の解析のことも考えて500回分試行していますが、このあと実は100回分しか使いません。
FileUtils.mkdir_p("Signature/nmf")
Parallel.each(1..500, in_processes: PCORE, progress: "NMF") do |seed|
decomposer = Rumale::Decomposition::NMF.new(n_components: 30, max_iter: 500, random_seed: seed)
decomposer.fit_transform(catalog)
File.binwrite("Signature/nmf/decomposer_#{seed}.dat", Marshal.dump(decomposer))
end
NMF: |=========================================================================|
1..500
これでデータ準備編はおしまいです。つぎに〜クラスタリング&可視化編〜にうつります。
🎨 〜クラスタリング&可視化編〜
🐳 目次
準備
- 🔖 セーブポイント用関数を準備する
- 📖 ライブラリを読み込む
データローディング
- 📂 シグネチャーのインデックスラベルを読み込む
- 📂 NArrayのNMF結果を読み込む
可視化してみる
- 📊 NMF結果のヒートマップを表示する
- 🔧 標準化してみる
- GaussianMixtureというのでクラスタリングする
- 📊 ヒートマップを描きなおす
t-SNE編
- 📚 Rumaleのt-SNEの使い方を確認する
- 🔧 t-SNEを実行する
- 💾 t-SNEの結果を保存する
- 📊 t-sneの結果を可視化する
- 📊 クラスタリングもふくめて可視化する
シグネチャー表示
- 📊 シグネチャー候補の可視化
そのほか
- 💾 CSVファイルを書き出す
1. 🔖 セーブポイント用関数を準備する
## 今回は Signature ディレクトリにオブジェクトを保存していく
require 'fileutils'
FileUtils.mkdir_p("Signature")
def sync(str, obj)
begin
if obj.nil?
Marshal.load(File.binread("Signature/#{str}.dat"))
else
File.binwrite("Signature/#{str}.dat", Marshal.dump(obj))
obj
end
rescue
Marshal.load(File.binread("Signature/#{str}.dat"))
end
end
2. 📖 ライブラリを読み込む
require 'numo/narray' # 行列計算
require 'numo/linalg' # OpenBLAS t-SNE高速化に重要
require 'iruby-plotly' # 可視化
require 'rumale' # 機械学習
3. 📂 シグネチャーのインデックスラベルを読み込む
signature_index = Marshal.load(File.binread("Signature/signature_index.dat"))
4. 📂 NArrayのNMF結果を読み込む
decomposer = (1..100).map do |i|
Marshal.load(File.binread("Signature/nmf/decomposer_#{i}.dat"))
end
components = Numo::DFloat.vstack(decomposer.map(&:components))
puts components.inspect
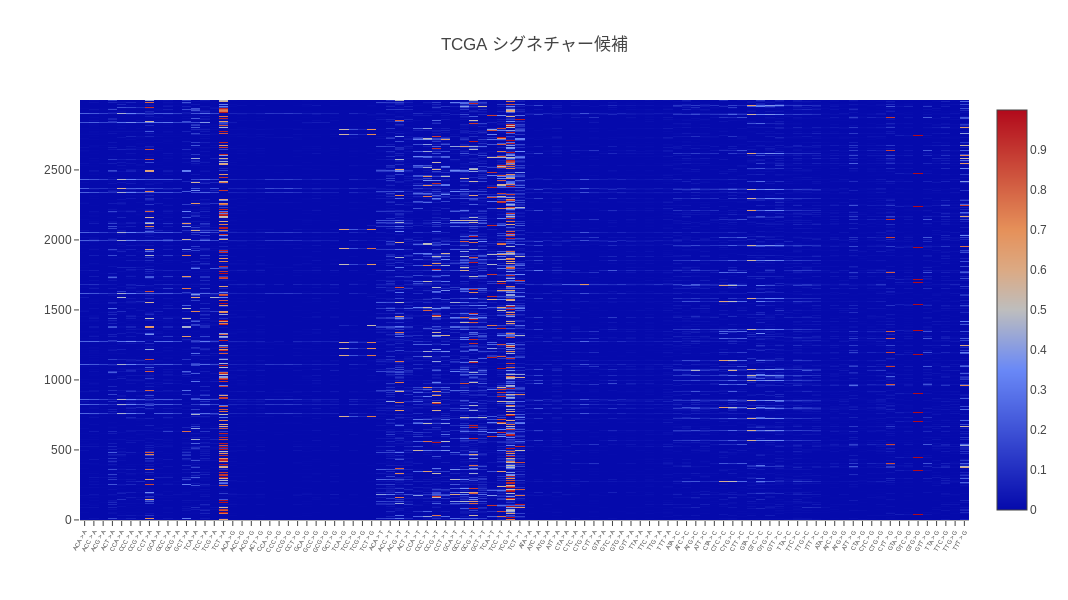
5. 📊 NMF結果のヒートマップを表示する
IRuby.plot(
{x: signature_index,
z: components.to_a,
type: "heatmap"},
{title: "TCGA シグネチャー候補",
xaxis: {tickangle: 300,
tickfont: {size: 6}},
height: 600,
width: 1000})
6. 🔧 標準化してみる
normalizer = Rumale::Preprocessing::StandardScaler.new
samples_std = normalizer.fit_transform(components.transpose)
samples_std = samples_std.transpose
puts samples_std.inspect
puts samples_std.var(axis: 1).minmax
[0.9999999999999953, 1.0000000000000049]
7. 🔧 GaussianMixtureというのでクラスタリングする
- KMeansはサンプルを等分しようとするので実データでは失敗します。
- 複数のシグネチャーが同じぐらい出現するという条件でシミュレーションを実行するとKMeansでもうまくいきます
- こちらのアルゴリズムの内容もよくわかりません…
gmm = Rumale::Clustering::GaussianMixture.new(n_clusters: 30, random_seed: 5)
pca = Rumale::Decomposition::PCA.new(n_components: 50)
samples_std_pca = pca.fit_transform(samples_std)
cluster_ids = gmm.fit_predict(samples_std_pca)
puts cluster_ids.inspect
Numo::Int32#shape=[3000]
[5, 9, 21, 29, 12, 7, 14, 24, 20, 27, 2, 11, 20, 25, 18, 29, 3, 18, 22, 7, ...]
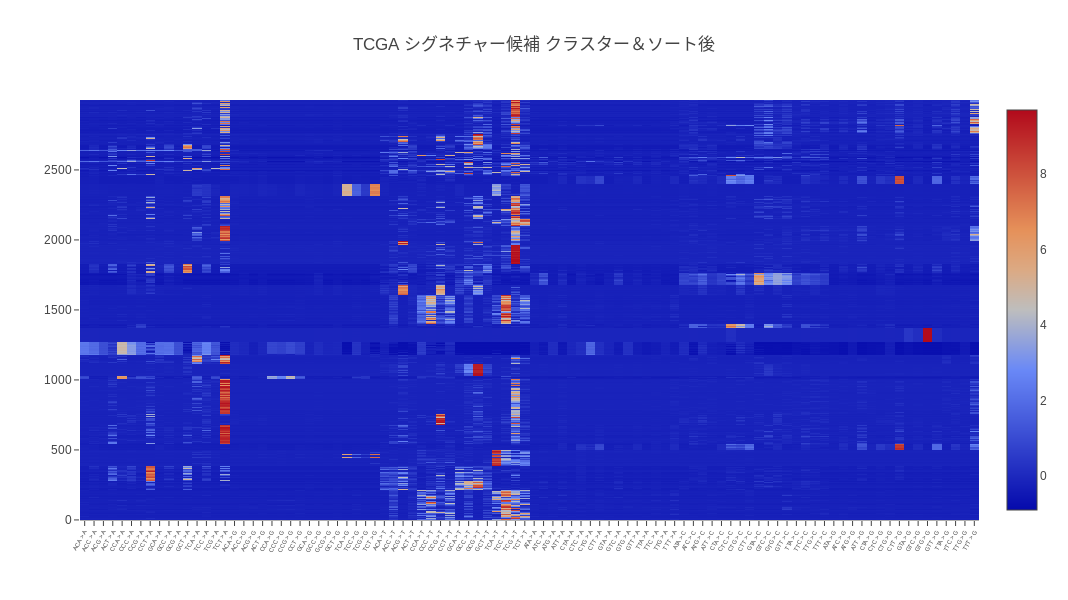
8. 📊 ヒートマップを描きなおす
IRuby.plot(
{x: signature_index,
z: samples_std[cluster_ids.sort_index, true].to_a,
type: "heatmap"},
{title: "TCGA シグネチャー候補 クラスター&ソート後",
xaxis: {tickangle: 300,
tickfont: {size: 6}},
height: 600,
width: 1000}
)
9. 📚 Rumaleのt-SNEの使い方を確認する
ri Rumale::Manifold::TSNE.new
⏰ t-SNEは時間がかかるので、何回か実行した時に時間を記録に残せるようにしておく
times = []
10. 🔧 t-SNEを実行する
start_time = Time.now
tsne = Rumale::Manifold::TSNE.new(n_components: 2, perplexity: 40.0, max_iter: 500, random_seed: 1)
tsne_representations = tsne.fit_transform(samples_std_pca)
times << Time.now - start_time
11. 💾 t-SNEの結果を保存する
tsne_representations = sync("tsne_representations", tsne_representations)
0
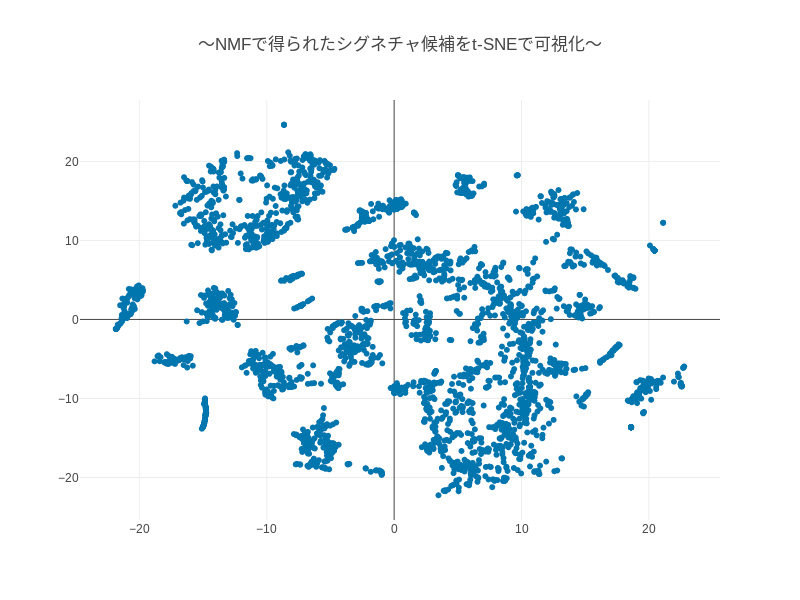
12. 📊 t-sneの結果を可視化する
x = tsne_representations[true, 0]
y = tsne_representations[true, 1]
IRuby.plot(x: x.to_a,
y: y.to_a,
mode: 'markers',
type: "scatter",
title: "〜NMFで得られたシグネチャ候補をt-SNEで可視化〜",
width: 800,
height: 600)
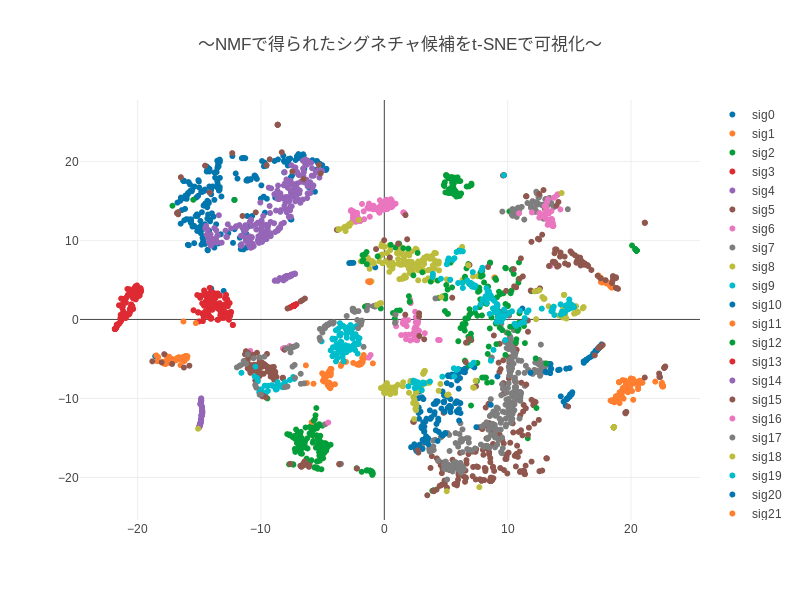
13. 📊 クラスタリングもふくめて可視化する
data = cluster_ids.to_a.uniq.sort.map do |l|
{
x: x[cluster_ids.eq(l)].to_a,
y: y[cluster_ids.eq(l)].to_a,
type: "scatter",
mode: 'markers',
name: "sig#{l}"
}
end
IRuby.plot(data, {title: "〜NMFで得られたシグネチャ候補をt-SNEで可視化〜", height: 600, width: 800})
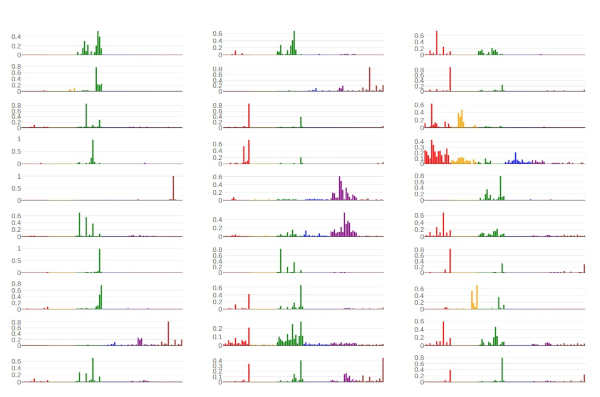
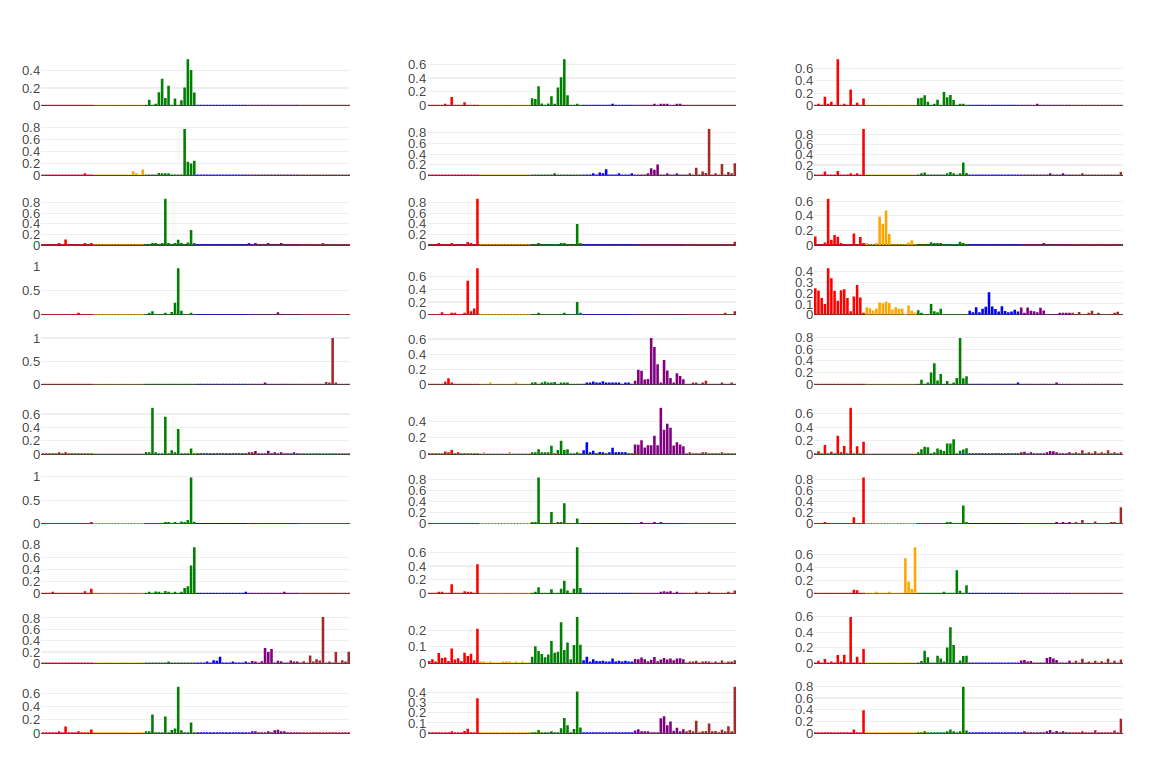
14. 🎨 シグネチャー候補の可視化
data = 30.times.map do |l|
s2 = components[cluster_ids.eq(l).where, true]
s = s2.mean(0)
color = %w(red orange green blue purple brown).map{|i| [i] * 16}.flatten
{y: s.to_a,
x: signature_index,
xaxis: "x#{l+1}",
yaxis: "y#{l+1}",
marker: {color: color},
type: "bar"}
end
layout = {
grid: {rows: 10, columns: 3,
pattern: 'independent'}}
(1..30).each do |i|
layout["xaxis#{i}".to_sym] = {showticklabels: false}
end
IRuby.plot(data, layout)
15. 💾 CSVファイルを書き出す
- 今回はRubyで頑張りましたが、🍊 Orange などのツールを使うとGUIで便利に解析することもできます。
- 他のツールでも何かできるように、CSVファイルで書き出します。
require 'csv'
CSV.open("Signature/nmf/signatures.csv", "w") do |csv|
csv << signature_index
samples_std.to_a.each do |ar|
csv << ar
end
end
0
結果
ここまでスクロールしてくれてありがとうございます。
今回は、GaussianMixtureというアルゴリズムをつかってクラスタリングを行うという単純な方法を採用しています。結果ですが、
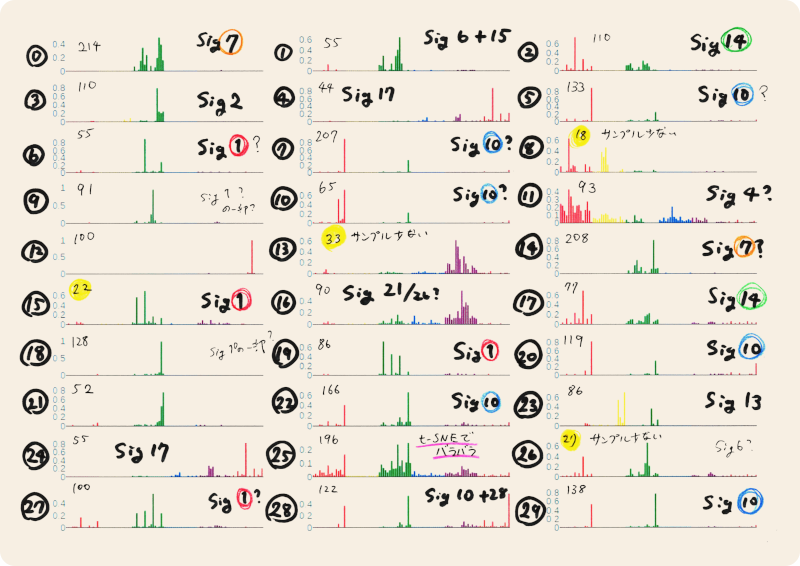
Signature1, Signature2, Signature6, Signature7, Signature10, Signature13, Signature14, Signature17, Signature21, Signature28 あたりは検出できた気がします。Signature4 は少し怪しいですが、検出できたことにします。
Sig1やSig10が何個も検出されています。これらの重複して検出されたSignatureはt-SNE で似たような位置にあることがわかります。候補8や候補13は、サンプル数が少ないのでNMFで誤検出された局所解かもしれません。
候補12や候補21は、少なくない回数検出されているにもかかわらずCosmicに似たようなSignatureが登録されていないものです。おそらく何らかのミスかアーチファクトだと思いますが、よくわかりません。エクソンのみの変異で本当に結果が出るか心配でしたが、思った以上にたくさんのシグネチャーが検出できました。
今回ある程度検出に成功したシグネチャー
| COSMIC signature | 関連する発がん要因 | 主ながん種 |
|---|---|---|
| Signature1 | 5-methylcytosineの脱メチル化, 加齢 | 広く認められる |
| Signature2 | APOBEC酵素活性 | 腺がんを中心に広くみられる |
| Signature4 | 喫煙 | 肺がん・頭頚部がん・肝臓がんなど |
| Signature6 | DNAミスマッチ修復異常 | 悪性リンパ腫など |
| Signature7 | 紫外線 | メラノーマ |
| Signature10 | POLE酵素異常 | 大腸がん・子宮がんなど |
| Signature13 | APOBEC酵素活性 | 腺がんを中心に広くみられる |
| Signature14 | POLE変異とミスマッチ修復異常 | 悪性リンパ腫など |
| Signature17 | 不明 | 食道がん・胃がんなど |
| Signature21 | 不明 | 胃がんなど |
| Signature28 | 不明 | 大腸がん・子宮がんなど |
おまけ 🍊 〜Orangeでもう少し詳しく結果をみてみよう〜
Orangeはスロベニアのリュブリャナ大学が開発しているGUIでデータ解析や機械学習ができるツールです。Jupyterや生Pythonを覚えなくてもクリック操作で解析ができてしまうという、本当に素晴らしいツールです。NMFも拡張機能を使用すれば実行できるようで、APIを叩くところを別にすれば、このページで紹介した処理はOrangeでみんな実行できてしまいそうです。ここでは後編の「クラスタリング&可視化編」をOrangeでやってみることにします。
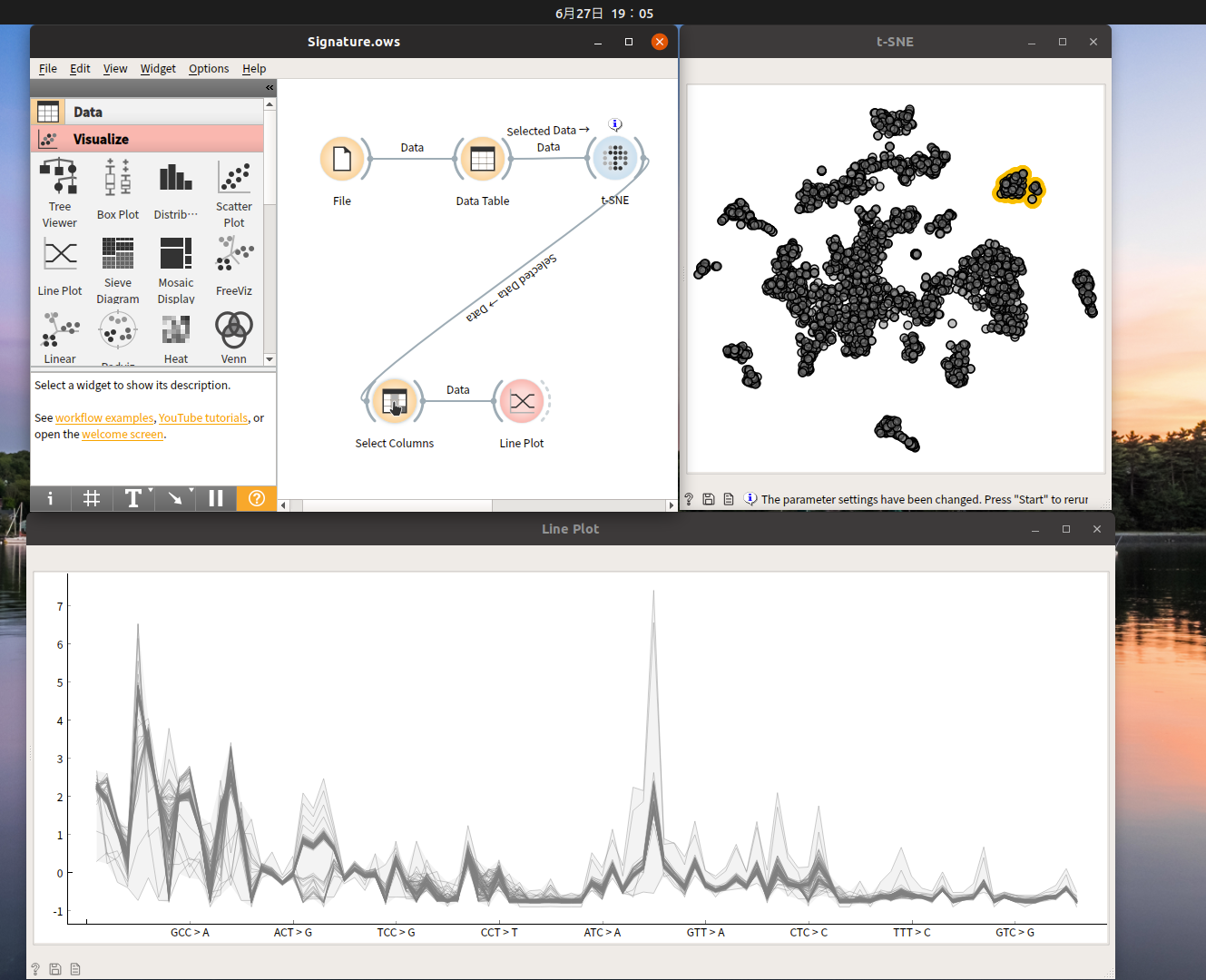
スクリーンショットのようにワークフローを定義して実行します。はい、便利すぎですよ (ToT) 💦
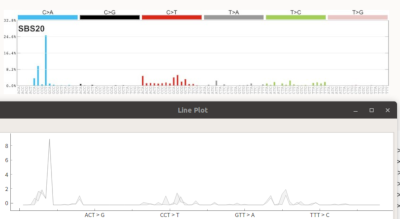
右のt-SNEのウィンドウで観察したいサンプルを選択すると、自動的に下のウィンドウでシグネチャーを描いています。
今描かれているのはSignature4だと思われます。

さらに細かく見ると、ここの小さい集団はSignature8に近いみたいだということがわかりました。
全体的にSignature10の影響が非常に強いです。Signature6や、Signature23は、それに近い候補が存在するが、Signature10番が強烈すぎて混ざってしまって正しく検出できてないのでは?という印象を受けました。
ごく小数ながらSignature20番に近いパターンが検出されていたりします。しかしせいぜい2個〜4個の候補が20番にちょっと似ている風だっただけなので偶然にすぎないかもしれません。
また候補13番はサンプル数が少ないと思っていましたが、単なる雑音として無視できなぐらい再現性を持って検出されるようです。うーん、これは一体何でしょう。どこか私のミスが隠れているのでしょうか。
Orangeはあまり注目されていませんがなぜでしょうか。それは解析APIを提供することでデータセンターに情報を集積するという大手ITプラットフォーマーの企業戦略と合致しないためかもしれません。ツールの良し悪しを判断するのは機械学習エンジニアです。しかし、付加価値を高める競争に日々晒されているであろうエンジニアからみると、Orangeは中途半端な機能しか持たない使えないツールに見えるかもしれません。あるいは逆に、それまでエンジニアが職人技でコードを書いて実現してきたプロセスの一部を、ボタンのクリックを数回やるだけで実行できるようにしているという意味で、機械学習職人の価値を毀損するツールのようにも見えて、反発したくなる心理が生じるかもしれません。ご心配なく。そういう心理は、エンジニアの世界に限らずどこのでもよくあることです。
しかし、これから「非エンジニア」の人々が使うデータ解析ツールは、PythonやJuliaではなく、Excel+Orangeのようなものになるんじゃないかなあと。個人的にはそんな気がちょっとしたのでした。
🎨 おまけ 〜扉絵の作り方〜
ペーパータオルを用意します 1

ぶっちゃけちゃんとした画用紙がある場合はそっちの方がいいです。
ボールペンで下絵を書いて、色鉛筆で塗ります。

スキャナーで画像を取り込みます。
↓スキャナーだとシワが写り込んでしまうので、カメラの方がいいかも

Gimpで画像を開いて、レベル補正など適当に一通りやったら、linear invert して Hue-Chroma で色合いを調整します。これでほとんど完成です。あとはお好みで symmetric nearest neighbor など気に入ったフィルターをいくつか追加します。
何のロゴかわかるかな?
この記事を書くにあたってお世話になったツールやサイトのロゴです。







左から順番に、Ruby, Rumale, Plotly, cBioPortal, COSMIC, Orange, GIMP でした。
この記事は以上です。
-
コンフォート サービスタオル200 https://pro.crecia.co.jp/product/search/index.php/item?id=11007 ↩