前回の記事では各ネットワークでFitnessを求め、150体いる全てのネットワークから最も成績が良いネットワークを探しました。
今回はこのgeneration=0のネットワークから子供世代(generation=1)を作ります。
交配と突然変異(④⑤)
次のコードを用意します。
import sys
import math
import random
from random import choice
elitism = 2
survival_threshold = 0.2
min_species_size = 2
species_fitness_func = max
class DefaultReproduction(object):
def __init__(self):
self.genome_indexer = count(1)
@staticmethod
def compute_spawn(adjusted_fitness, previous_sizes, pop_size, min_species_size):
af_sum = sum(adjusted_fitness)
spawn_amounts = []
for af, ps in zip(adjusted_fitness, previous_sizes):
if af_sum > 0:
s = max(min_species_size, af / af_sum * pop_size)
else:
s = min_species_size
d = (s - ps) * 0.5
c = int(round(d))
spawn = ps
if abs(c) > 0:
spawn += c
elif d > 0:
spawn += 1
elif d < 0:
spawn -= 1
spawn_amounts.append(spawn)
total_spawn = sum(spawn_amounts)
norm = pop_size / total_spawn
spawn_amounts = [max(min_species_size, int(round(n * norm))) for n in spawn_amounts]
return spawn_amounts
def configure_crossover(self, genome1, genome2, child):
if genome1.fitness > genome2.fitness:
parent1, parent2 = genome1, genome2
else:
parent1, parent2 = genome2, genome1
for key, cg1 in parent1.connections.items():
cg2 = parent2.connections.get(key)
if cg2 is None:
child.connections[key] = cg1.copy()
else:
child.connections[key] = cg1.crossover(cg2)
parent1_set = parent1.nodes
parent2_set = parent2.nodes
for key, ng1 in parent1_set.items():
ng2 = parent2_set.get(key)
assert key not in child.nodes
if ng2 is None:
child.nodes[key] = ng1.copy()
else:
child.nodes[key] = ng1.crossover(ng2)
def update(self, species_set, generation):
species_data = []
for sid, s in species_set.species.items():
if s.fitness_history:
prev_fitness = max(s.fitness_history)
else:
prev_fitness = -sys.float_info.max
s.fitness = species_fitness_func(s.get_fitnesses())
s.fitness_history.append(s.fitness)
s.adjusted_fitness = None
if prev_fitness is None or s.fitness > prev_fitness:
s.last_improved = generation
species_data.append((sid, s))
species_data.sort(key=lambda x: x[1].fitness)
result = []
species_fitnesses = []
num_non_stagnant = len(species_data)
for idx, (sid, s) in enumerate(species_data):
result.append((sid, s))
species_fitnesses.append(s.fitness)
return result
def reproduce(self, species, pop_size, generation):
all_fitnesses = []
remaining_species = []
passing = None
for stag_sid, stag_s in self.update(species, generation):
all_fitnesses.extend(m.fitness for m in stag_s.members.values())
remaining_species.append(stag_s)
if not remaining_species:
species.species = {}
return {}
min_fitness = min(all_fitnesses)
max_fitness = max(all_fitnesses)
fitness_range = max(1.0, max_fitness - min_fitness)
for afs in remaining_species:
msf_values = list([m.fitness for m in afs.members.values()])
msf=sum(map(float, msf_values)) / len(msf_values)
af = (msf - min_fitness) / fitness_range
afs.adjusted_fitness = af
adjusted_fitnesses = [s.adjusted_fitness for s in remaining_species]
previous_sizes = [len(s.members) for s in remaining_species]
spawn_amounts = self.compute_spawn(adjusted_fitnesses, previous_sizes, pop_size, min_species_size)
new_population = {}
species.species = {}
for spawn, s in zip(spawn_amounts, remaining_species):
spawn = max(spawn, elitism)
assert spawn > 0
old_members = list(s.members.items())
s.members = {}
species.species[s.key] = s
old_members.sort(reverse=True, key=lambda x: x[1].fitness)
for i, m in old_members[:elitism]:
new_population[i] = m
spawn -= 1
if spawn <= 0:
continue
repro_cutoff = int(math.ceil(survival_threshold *
len(old_members)))
repro_cutoff = max(repro_cutoff, 2)
old_members = old_members[:repro_cutoff]
while spawn > 0:
parent1_id, parent1 = random.choice(old_members)
parent2_id, parent2 = random.choice(old_members)
gid = next(self.genome_indexer)
for a, b in list(new_population.items()):
if a == gid:
passing = True
if passing == True:
passing = None
continue
child = DefaultGenome(gid)
self.configure_crossover(parent1, parent2, child)
mutate = Mutate()
mutate.mutate(child)
new_population[gid] = child
spawn -= 1
return new_population
あとは突然変異を起こすコードも用意します。
class Mutate(object):
def __init__(self):
pass
def mutate(self,child):
from random import random
if random() < 0.2:#node_add_probability
self.mutate_add_node(child)
if random() < 0.2:#node_delete_probability
self.mutate_delete_node(child)
if random() < 0.5:#connection_add_probability
self.mutate_add_connection(child)
if random() < 0.5:#connection_delete_probability
self.mutate_delete_connection(child)
for cg in child.connections.values():
cg.mutate()
for ng in child.nodes.values():
ng.mutate()
def mutate_add_node(self,child):
if not child.connections:
return
conn_to_split = choice(list(child.connections.values()))
if child.node_indexer is None:
child.node_indexer = count(max(list(child.nodes)) + 1)
new_node_id = next(child.node_indexer)
ng = DefaultGenome.create_node( new_node_id)
child.nodes[new_node_id] = ng
conn_to_split.enabled = False
i, o = conn_to_split.key
self.add_connection(child,i, new_node_id, 1.0, True)
self.add_connection(child,new_node_id, o, conn_to_split.weight, True)
def add_connection(self, child,input_key, output_key, weight, enabled):
key = (input_key, output_key)
connection = DefaultConnectionGene(key)
connection.init_attributes()
connection.weight = weight
connection.enabled = enabled
child.connections[key] = connection
def mutate_add_connection(self,child):
possible_outputs = list(child.nodes)
out_node = choice(possible_outputs)
input_keys=[-i - 1 for i in range(num_inputs)]
possible_inputs = possible_outputs + input_keys
in_node = choice(possible_inputs)
key = (in_node, out_node)
if key in child.connections:
return
if in_node == out_node:
return
for a, b in list(child.connections):
if out_node == a:
if in_node == b:
return
cg = DefaultGenome.create_connection( in_node, out_node)
child.connections[cg.key] = cg
def mutate_delete_node(self,child):
output_keys = [i for i in range(num_outputs)]
available_nodes = [k for k in child.nodes if k not in output_keys]
if not available_nodes:
return -1
del_key = choice(available_nodes)
connections_to_delete = set()
for k, v in child.connections.items():
if del_key in v.key:
connections_to_delete.add(v.key)
for key in connections_to_delete:
del child.connections[key]
del child.nodes[del_key]
return del_key
def mutate_delete_connection(self,child):
if child.connections:
key = choice(list(child.connections.keys()))
del child.connections[key]
ここで定義されたDefaultReproductionは交配に使われるメソッドをもちます。
popus = DefaultReproduction()
population = popus.reproduce(species, pop_size, generation)
最初に1から始まっていないのはgeneration=0の成績が最もよかった二つのネットワークがそのまま次のgenerationに移ったからです。他のネットワークをみると、突然変異の影響でNodesが増えていたり、Connectionsが減っていたりする様子がわかります。
ここで交配の際に起こる、二つのネットワークのcrossoverについて説明します。
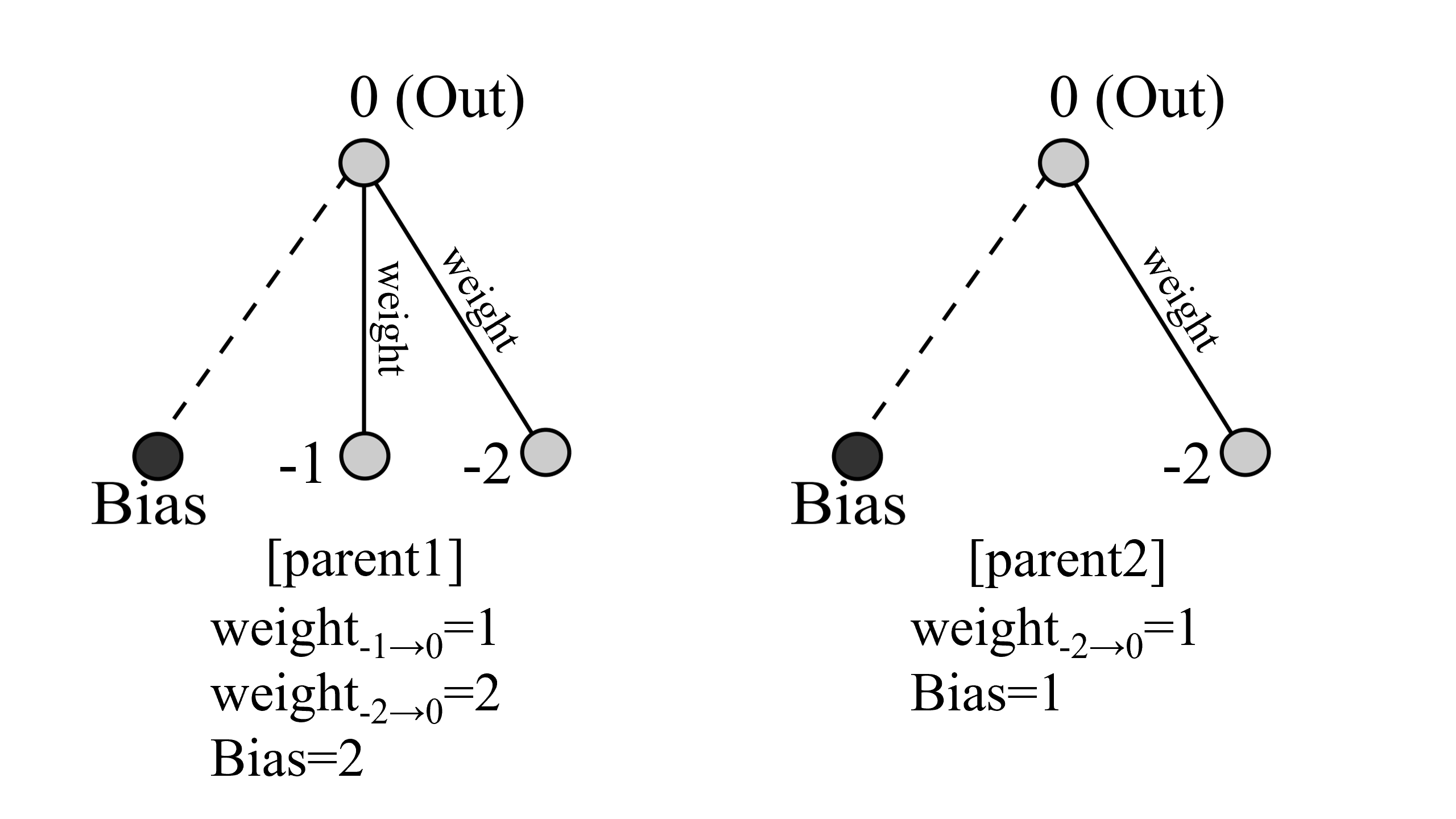
crossover
このようなparent1とparent2がいたとします。二つを比較して、NodesやConnectionsに違いがあれば、それらは子供に継承されます。この例だと、key=(-1,0)のConnectionsと-1のNodesが継承されます。
次に共通するNodesまたはConnectionsがあれば確率的(今回は1/2)にどちらかが選ばれます。
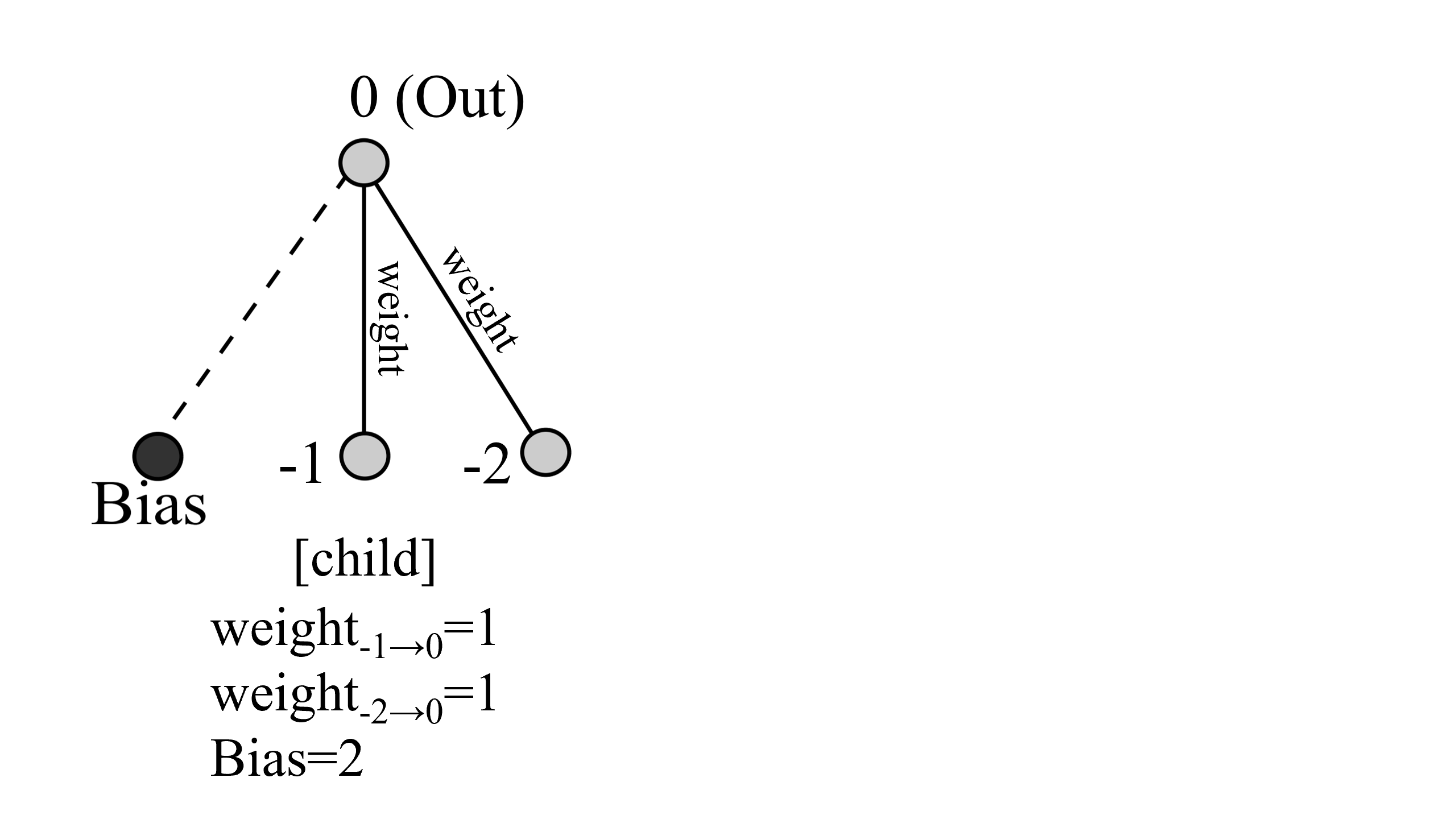
そしてcrossoverの結果、次のような子供が作られます。
このようにして任意で選ばれた二つのネットワークから子供が150体(種の種類が増えると僅かに前後します)作られます。
そして、generationを一つ繰り上げます。
generation += 1
次回は今までの操作をwhile文でループさせ、最終的な結果をみます。