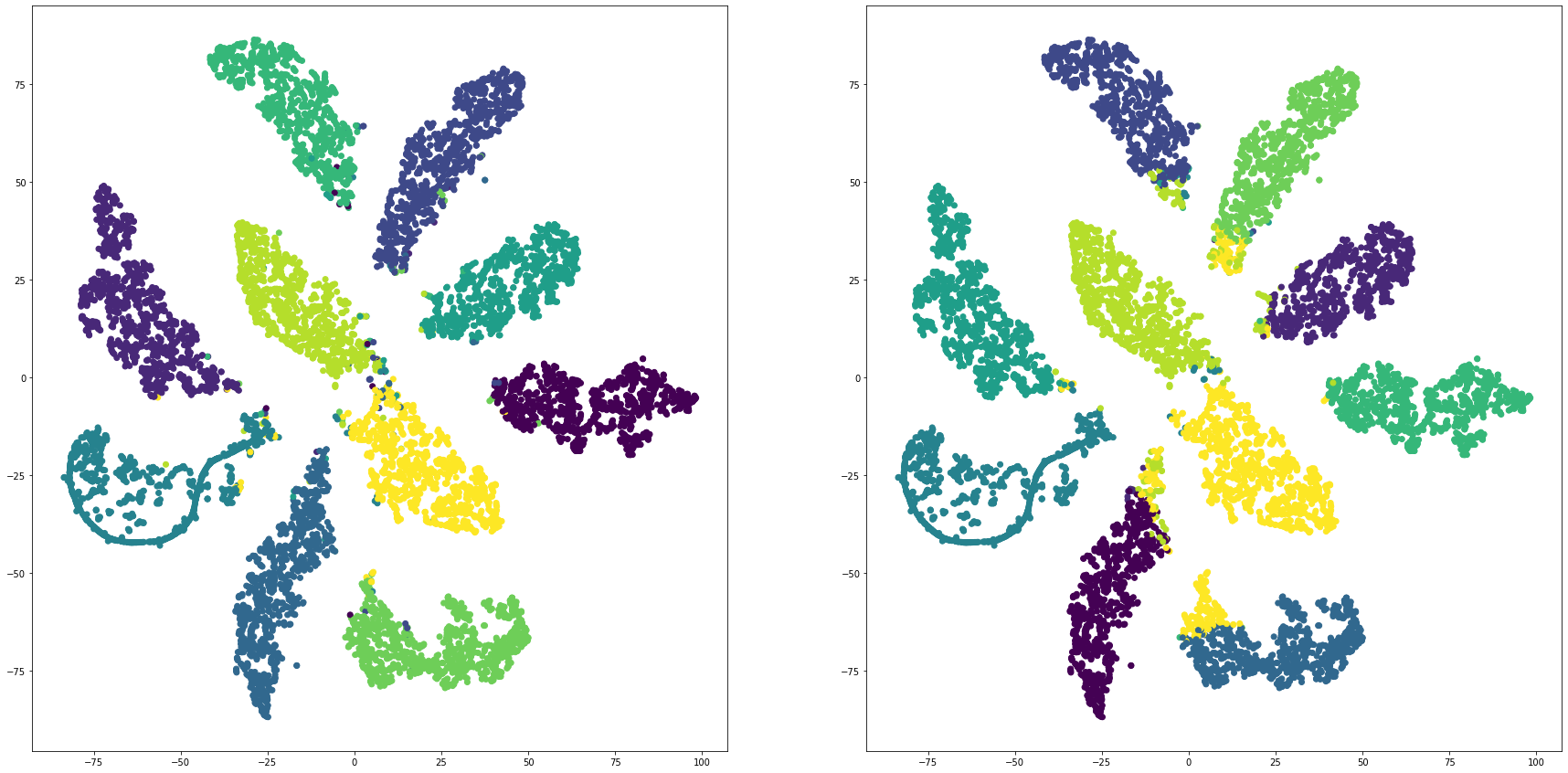
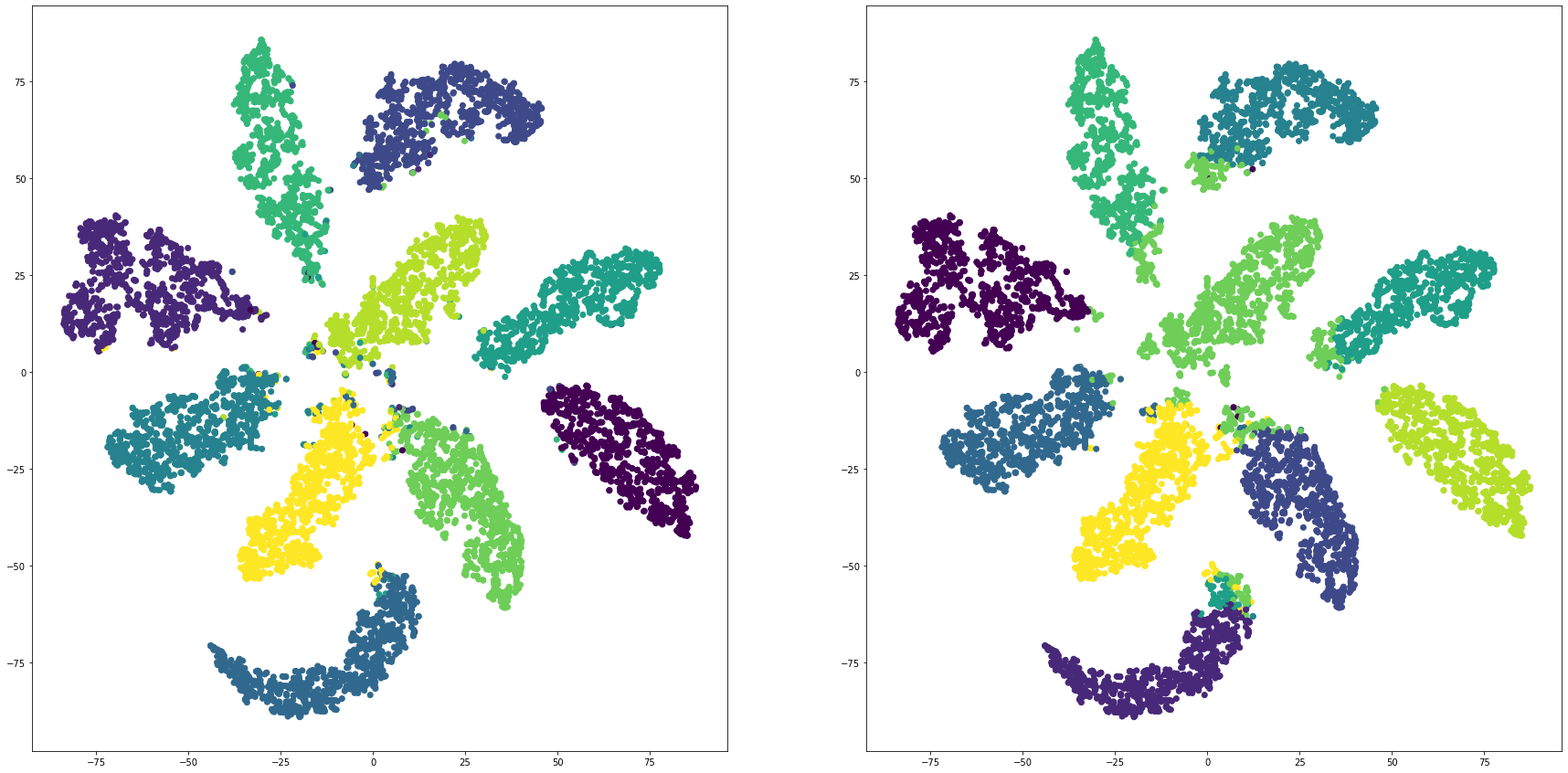
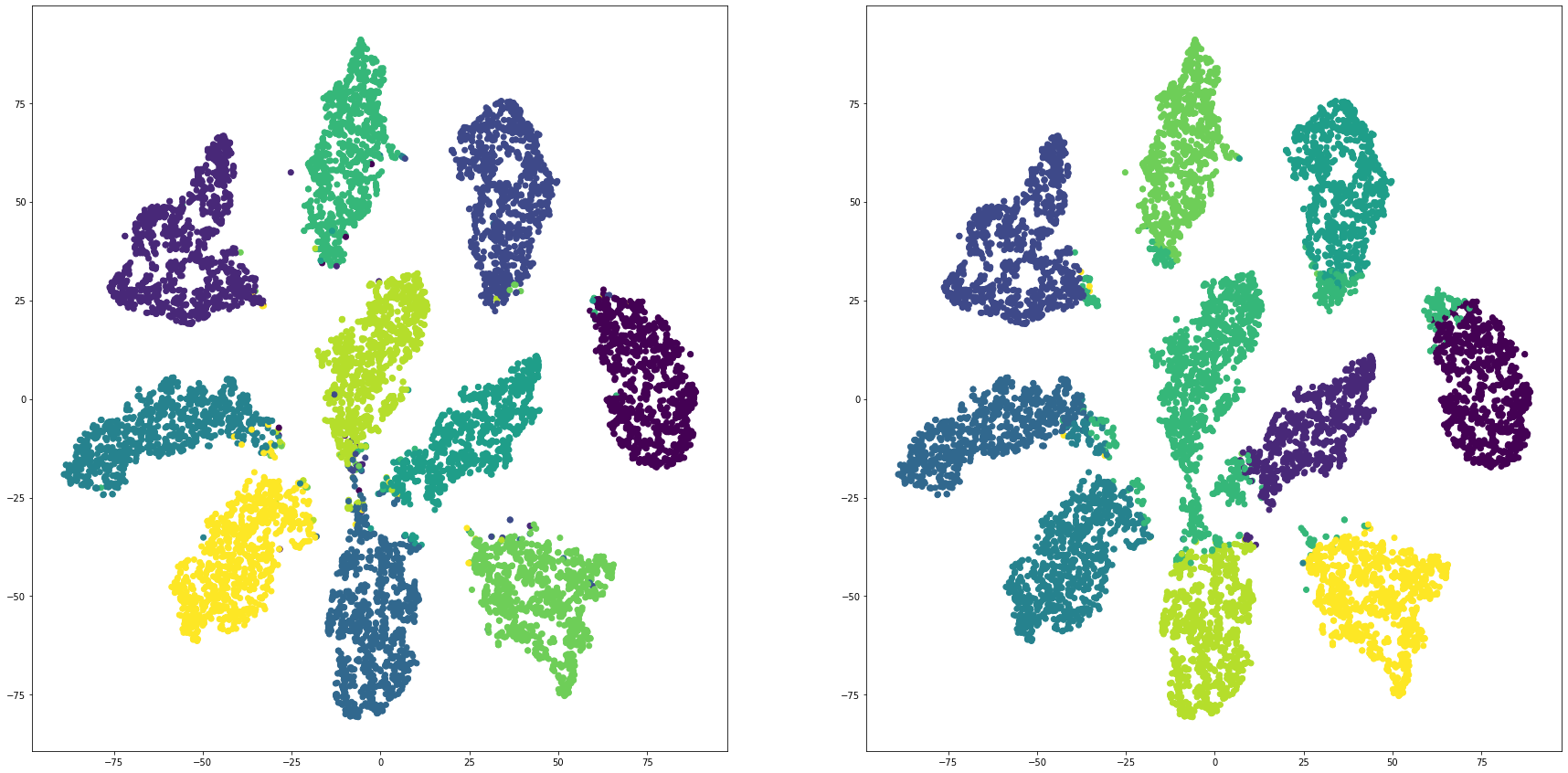
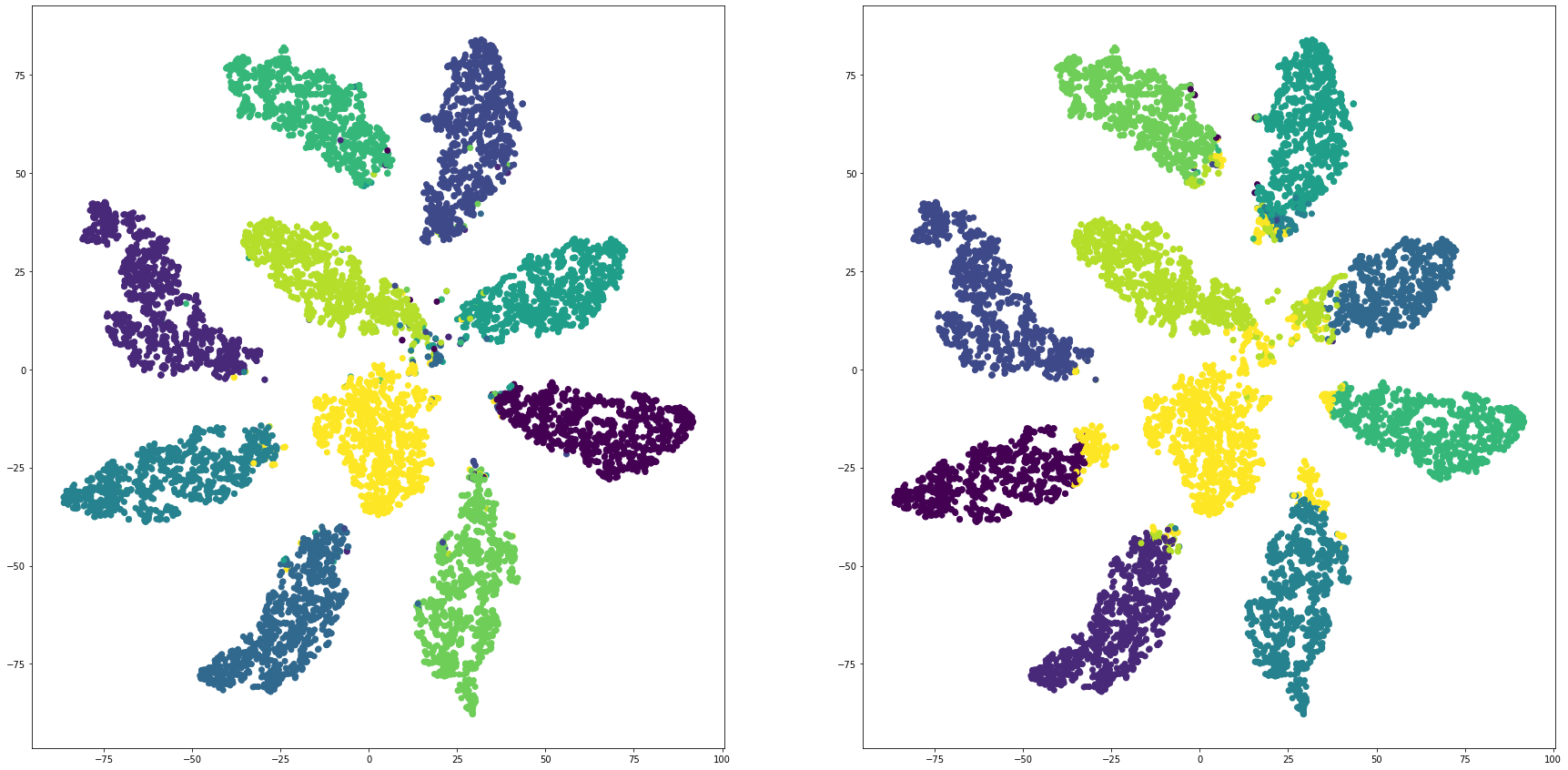
mnistを全結合層で教師あり学習し、最終段をクラスタリングして評価する
# 必要なライブラリのインポート
import keras
from keras.datasets import mnist
import numpy as np
import pandas as pd
import sklearn
# Jupyter notebookを利用している際に、notebook内にplot結果を表示するようにする
import matplotlib.pyplot as plt
%matplotlib inline
Using TensorFlow backend.
feature_dims = range(2, 12)
# Kerasの関数でデータの読み込み。データをシャッフルして学習データと訓練データに分割
(x_train, y_train), (x_test, y_test) = mnist.load_data()
# 2次元データを数値に変換
x_train = x_train.reshape(60000, 784)
x_test = x_test.reshape(10000, 784)
# 型変換
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# 255で割ったものを新たに変数とする
x_train /= 255
x_test /= 255
# one-hot encodingを施すためのメソッド
from keras.utils.np_utils import to_categorical
# クラス数は10
num_classes = 10
y_train = y_train.astype('int32')
y_test = y_test.astype('int32')
labels = y_test
# one-hot encoding
y_train = to_categorical(y_train, num_classes)
y_test = to_categorical(y_test, num_classes)
def fitting(feature_dim, x_train, y_train, x_test, y_test):
# 必要なライブラリのインポート、最適化手法はAdamを使う
from keras.models import Sequential
from keras.layers import Dense, Dropout
from keras.optimizers import Adam
import gc
# モデル作成
model = Sequential()
model.add(Dense(512, activation='relu', input_shape=(784,)))
model.add(Dropout(0.2))
model.add(Dense(512, activation='relu'))
model.add(Dropout(0.2))
model.add(Dense(feature_dim, activation='relu')) # 特徴量として取り出すための層を追加
model.add(Dense(10, activation='softmax'))
model.summary()
# バッチサイズ、エポック数
batch_size = 128
epochs = 20
model.compile(loss='categorical_crossentropy',
optimizer=Adam(),
metrics=['accuracy'])
history = model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(x_test, y_test))
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
# #Accuracy
# print(history.history.keys())
# plt.plot(history.history['accuracy'])
# plt.plot(history.history['val_accuracy'])
# plt.title('model accuracy')
# plt.ylabel('accuracy')
# plt.xlabel('epoch')
# plt.legend(['train', 'test'], loc='upper left')
# plt.show()
# #loss
# plt.plot(history.history['loss'])
# plt.plot(history.history['val_loss'])
# plt.title('model loss')
# plt.ylabel('loss')
# plt.xlabel('epoch')
# plt.legend(['train', 'test'], loc='upper left')
# plt.show()
model.pop() # 最終段のsoftmax層を取り除いて、特徴量の層を最終段とする
model.summary()
result = model.predict(x_test)
keras.backend.clear_session() # ←これです
gc.collect()
from IPython.display import clear_output
clear_output()
return (history, model, result)
# model = fitting(10, x_train, y_train, x_test, y_test)
models = [None] * len(feature_dims)
histories = [None] * len(feature_dims)
results = [None] * len(feature_dims)
for i in range(len(feature_dims)):
(histories[i], models[i], results[i]) = fitting(feature_dims[i], x_train, y_train, x_test, y_test)
# model.save('model/mnist-10')
# model = keras.models.load_model('model/mnist-10')
# for i in range(len(feature_dims)):
# models[i].pop() # 最終段のsoftmax層を取り除いて、特徴量の層を最終段とする
# models[i].summary()
# result = model.predict(x_test)
# results = [None] * len(feature_dims)
# for i in range(len(feature_dims)):
# keras.backend.clear_session()
# results[i] = models[i].predict(x_test)
def tsne(result):
#t-SNEで次元削減
from sklearn.manifold import TSNE
tsne = TSNE(n_components=2, random_state = 0, perplexity = 30, n_iter = 1000)
return tsne.fit_transform(result)
# tsne = tsne(result)
tsnes = [None] * len(feature_dims)
for i in range(len(feature_dims)):
tsnes[i] = tsne(results[i])
# df = pd.DataFrame(tsne, columns = ['x', 'y'])
# df['label'] = labels
def km(n_clusters, result):
# k-meansでクラスタリングする
from sklearn.cluster import KMeans
return KMeans(n_clusters).fit_predict(result)
# km = km(10, result)
# df['km'] = km
kms = [None] * len(feature_dims)
for i in range(len(feature_dims)):
kms[i] = km(10, results[i])
def DBSCAN(n_clusters, result):
from sklearn.cluster import DBSCAN
db = DBSCAN(eps=0.2, min_samples=n_clusters).fit(result)
return db.labels_
# dbscan = DBSCAN(20, result)
# df['DBSCAN'] = dbscan
def hierarchy(result):
from scipy.cluster.hierarchy import linkage, dendrogram
result1 = linkage(result,
metric = 'braycurtis',
#metric = 'canberra',
#metric = 'chebyshev',
#metric = 'cityblock',
#metric = 'correlation',
#metric = 'cosine',
#metric = 'euclidean',
#metric = 'hamming',
#metric = 'jaccard',
#method= 'single')
method = 'average')
#method= 'complete')
#method='weighted')
return result1
# hierarchy = hierarchy(result)
# display(hierarchy)
# def cluster_visualization(x, y, label, cluster, method, n_clusters):
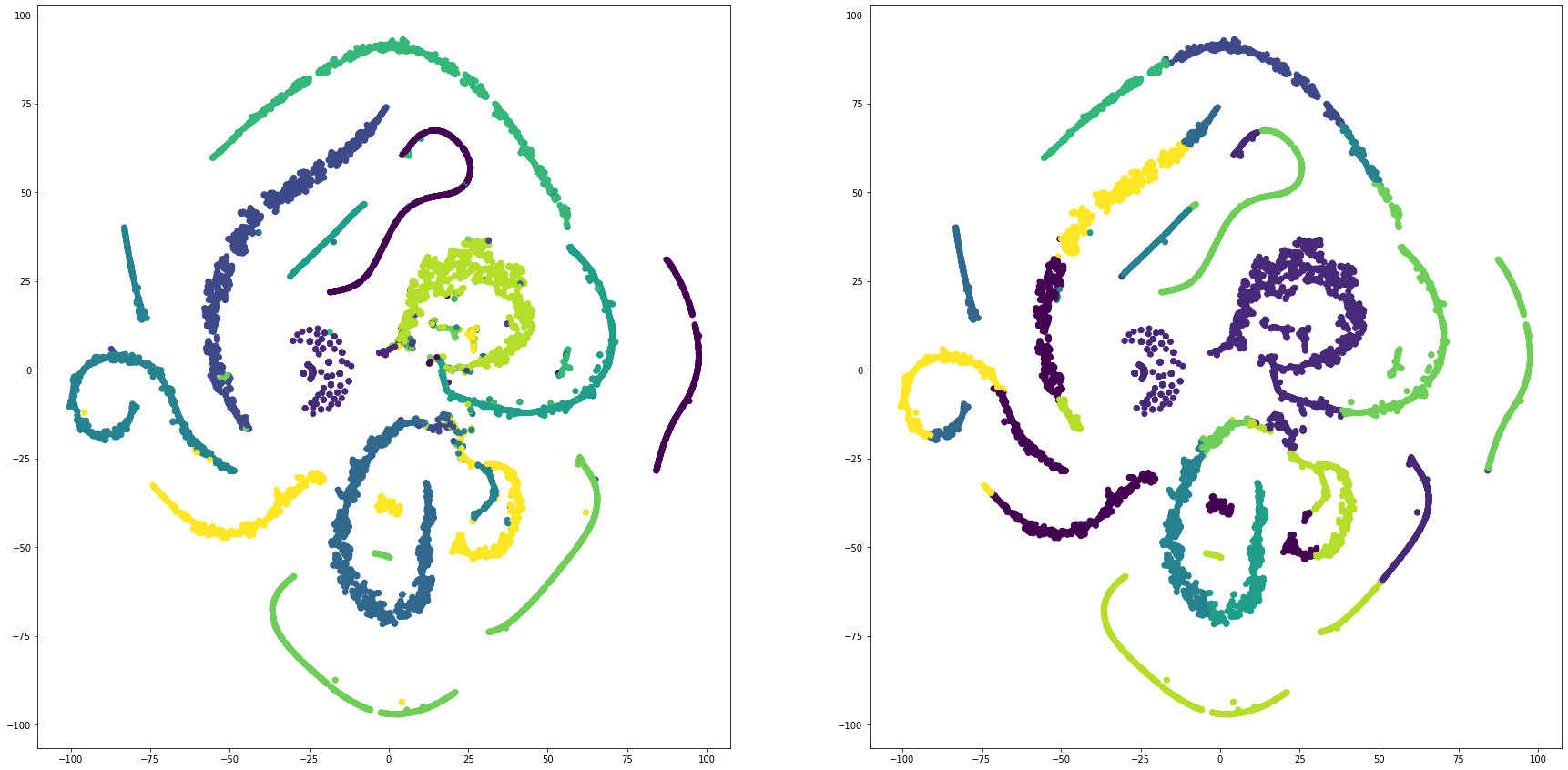
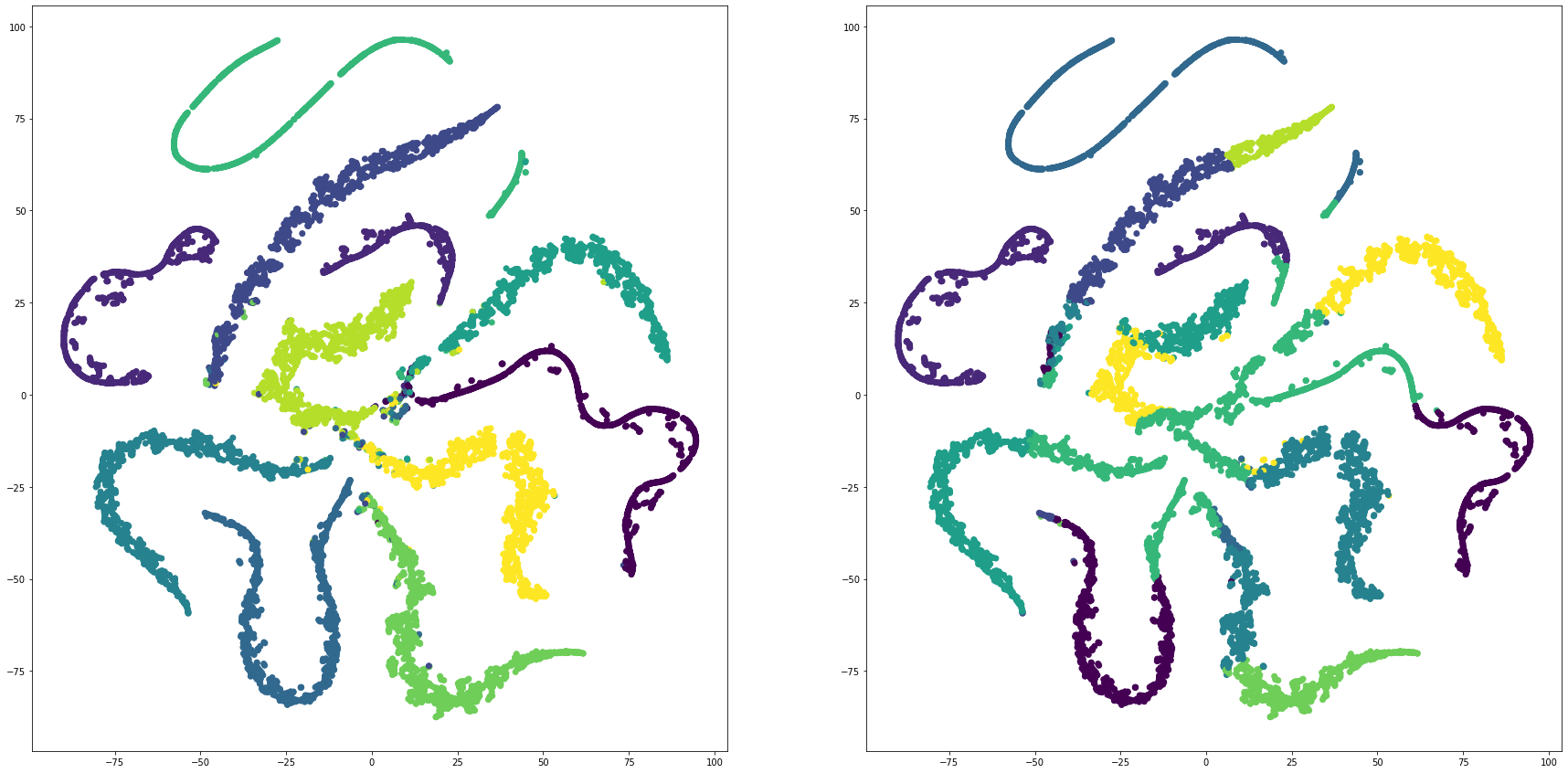
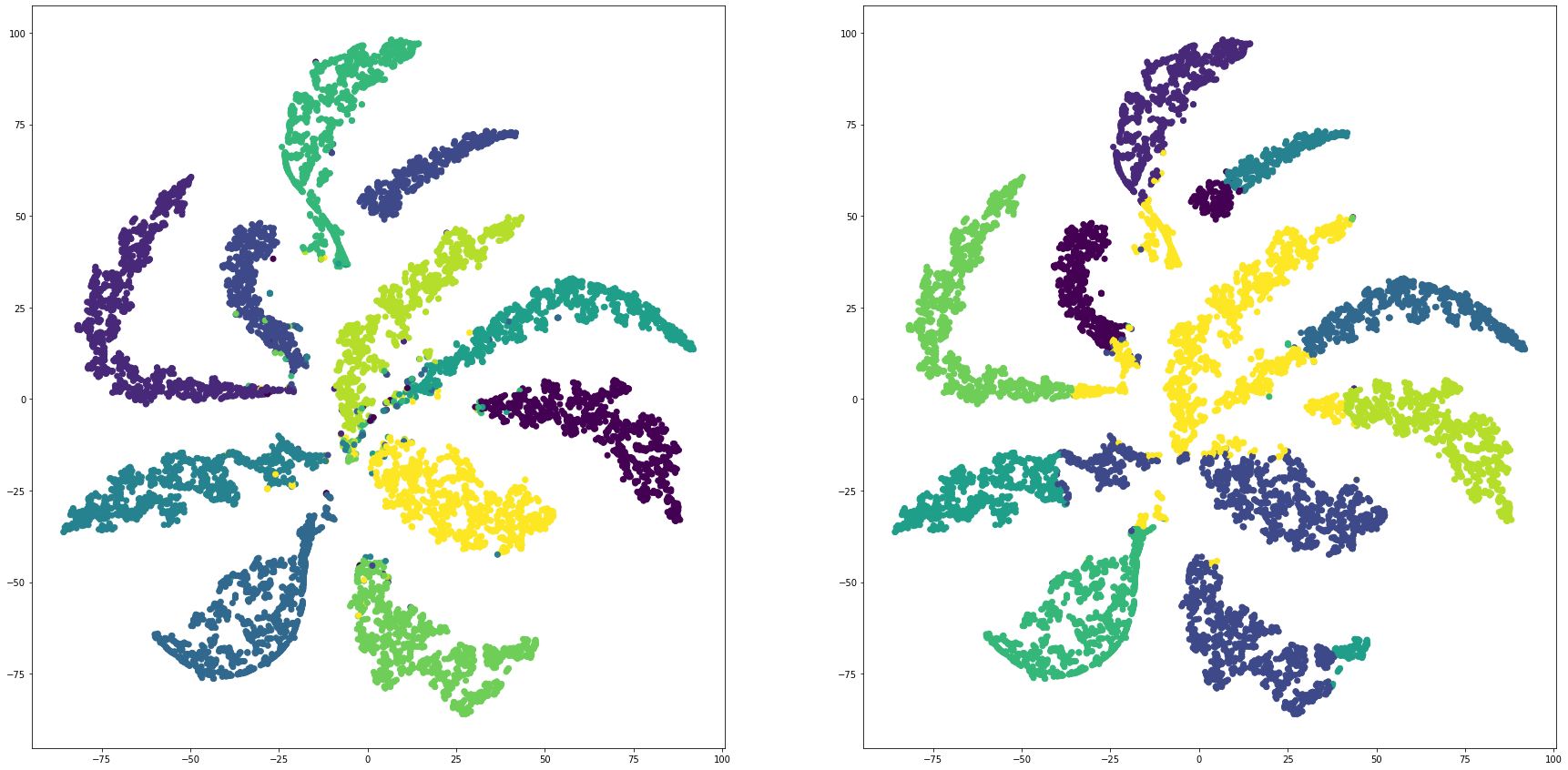
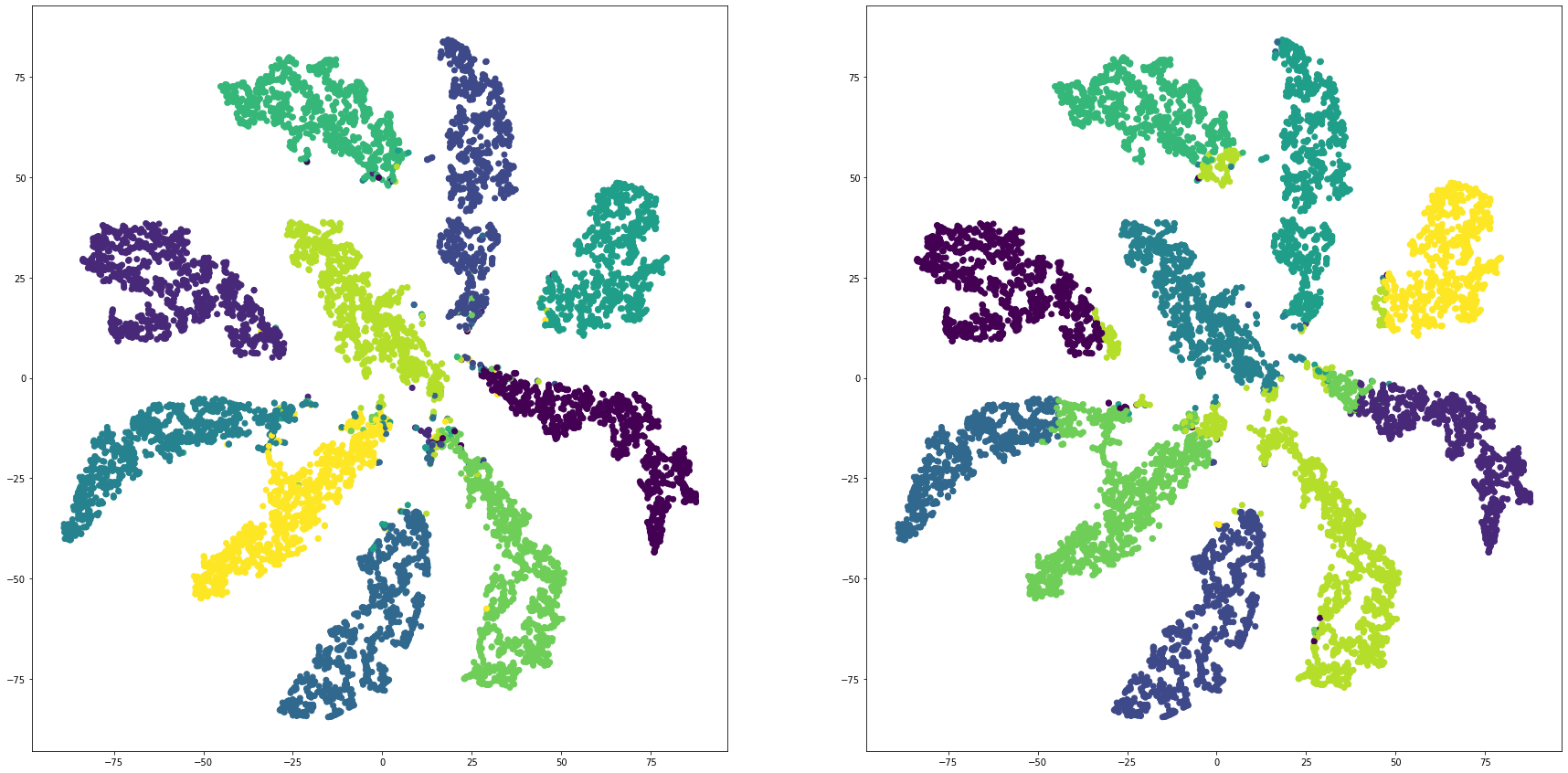
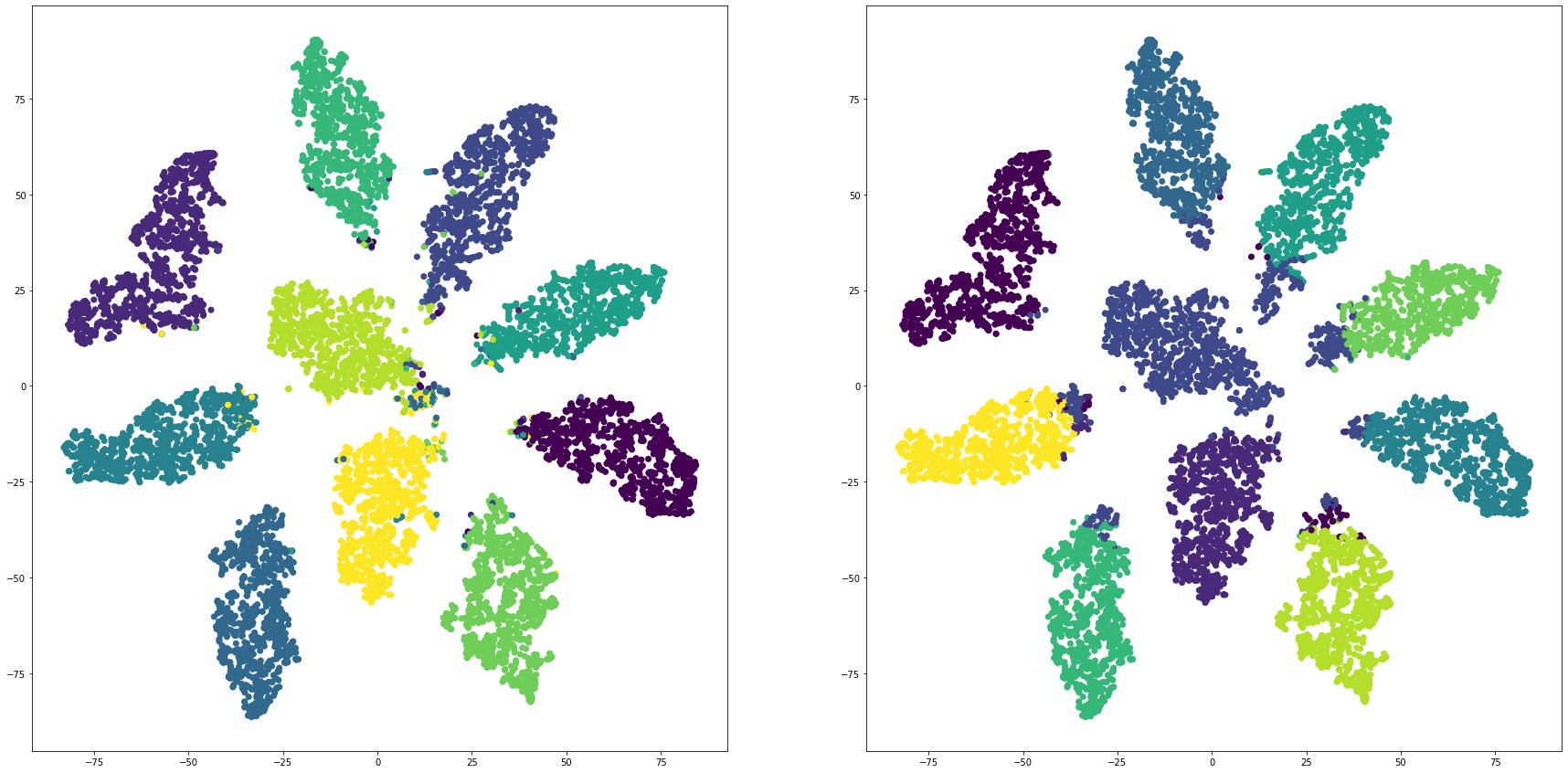
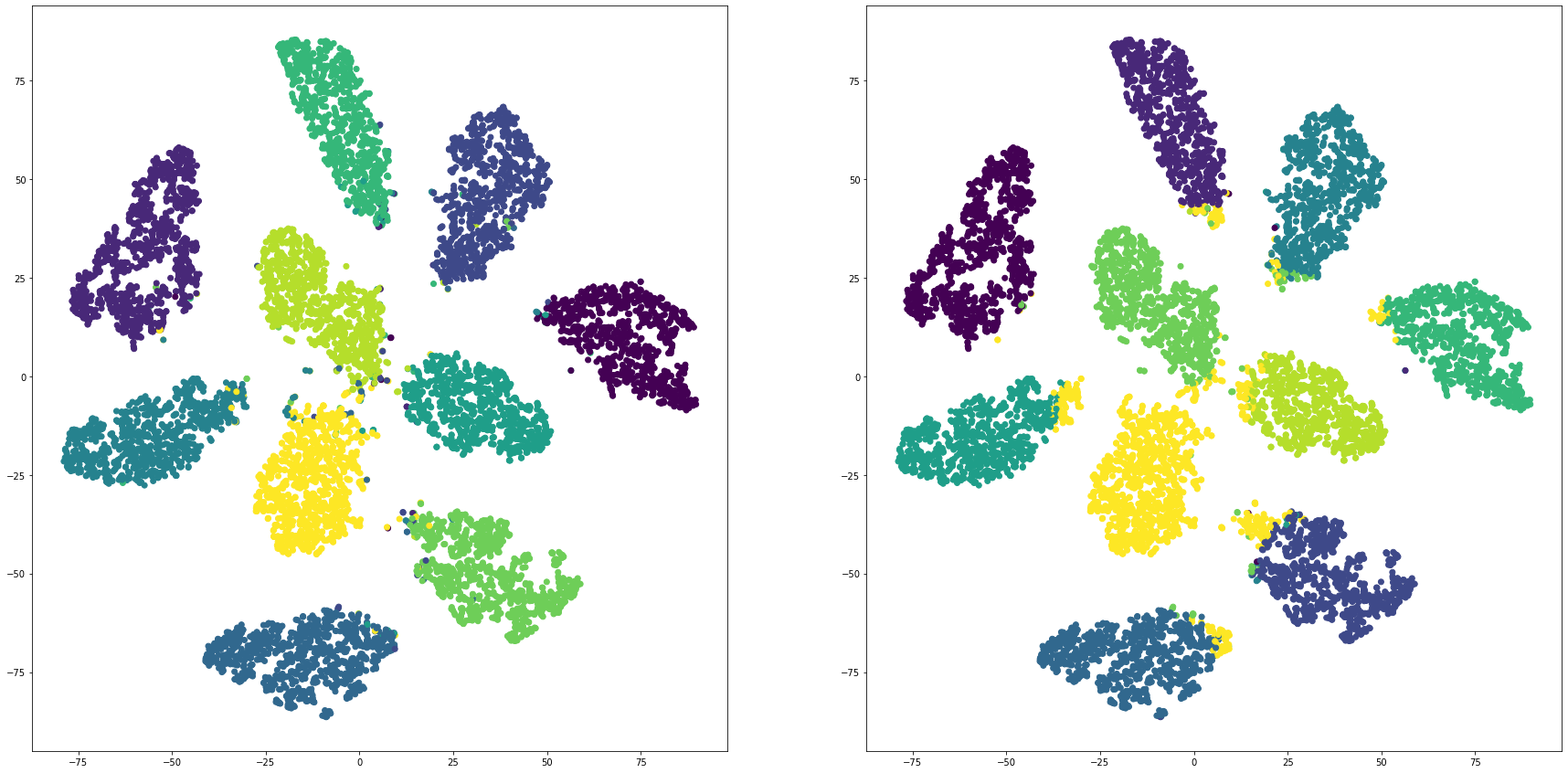
def cluster_visualization(x, y, label, cluster):
plt.figure(figsize = (30, 15))
plt.subplot(1,2,1)
plt.scatter(x, y, c=label)
# for i in range(10):
# tmp_df = df[df['label'] == i]
# plt.scatter(tmp_df['x'], tmp_df['y'], label=i)
# plt.legend(loc='upper left', bbox_to_anchor=(1,1))
plt.subplot(1,2,2)
plt.scatter(x, y, c=cluster)
# for i in range(n_clusters):
# tmp_df = df[df[method] == i]
# plt.scatter(tmp_df['x'], tmp_df['y'], label=i)
# plt.legend(loc='upper left', bbox_to_anchor=(1,1))
for i in range(len(feature_dims)):
cluster_visualization(tsnes[i][:,0], tsnes[i][:,1], labels, kms[i])
# https://qiita.com/mamika311/items/75c24f6892f85593f7e7
from sklearn.metrics.cluster import adjusted_rand_score
for i in range(len(feature_dims)):
print("dim:" + str(feature_dims[i]) + " ARI: " + str(adjusted_rand_score(labels, kms[i])))
dim:2 ARI: 0.36573507862590254
dim:3 ARI: 0.49974179932107105
dim:4 ARI: 0.6248257814760337
dim:5 ARI: 0.8225287029746797
dim:6 ARI: 0.8495039832620757
dim:7 ARI: 0.8417680081349097
dim:8 ARI: 0.8423268187793562
dim:9 ARI: 0.8450473012143238
dim:10 ARI: 0.836035505993697
dim:11 ARI: 0.8815919206871302
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.normalized_mutual_info_score.html
# https://qiita.com/kotap15/items/38289edfe822005e1e44
from sklearn.metrics import normalized_mutual_info_score
# display(normalized_mutual_info_score(labels, df['km']))
for i in range(len(feature_dims)):
print("dim:" + str(feature_dims[i]) + " NMI: " + str(normalized_mutual_info_score(labels, kms[i])))
dim:2 NMI: 0.5759443563915843
dim:3 NMI: 0.6735454178249051
dim:4 NMI: 0.7745736983918213
dim:5 NMI: 0.8626814016489588
dim:6 NMI: 0.8759626968874756
dim:7 NMI: 0.8766399602087444
dim:8 NMI: 0.8830520742914061
dim:9 NMI: 0.8706715369843739
dim:10 NMI: 0.8721342625213994
dim:11 NMI: 0.8992713472017846
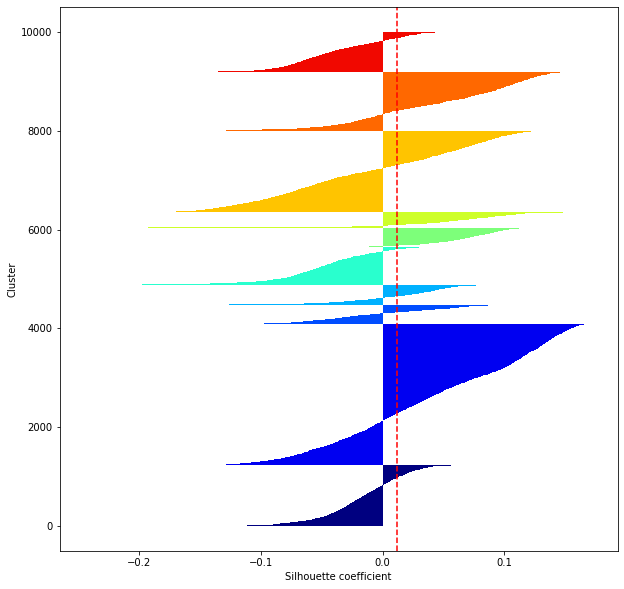
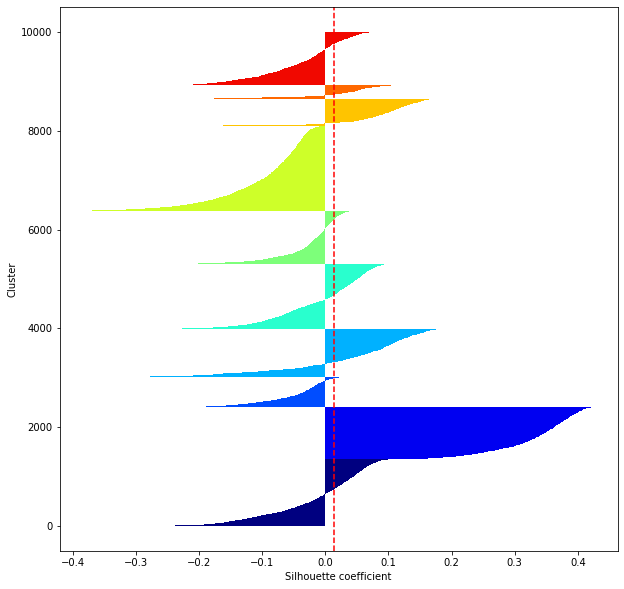
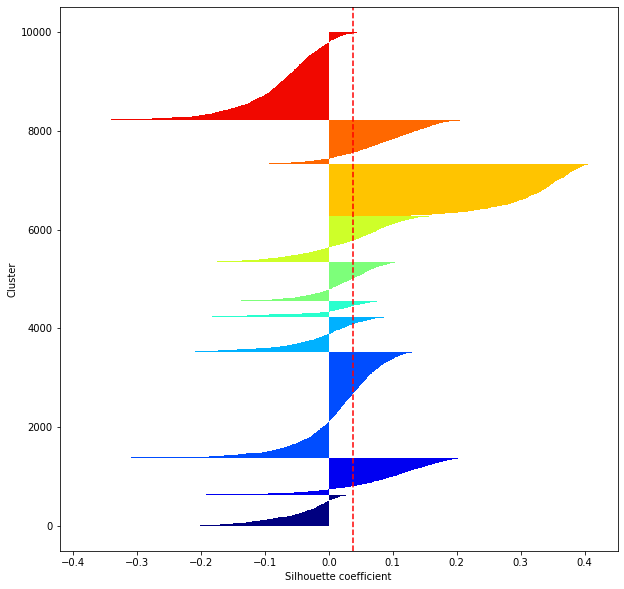
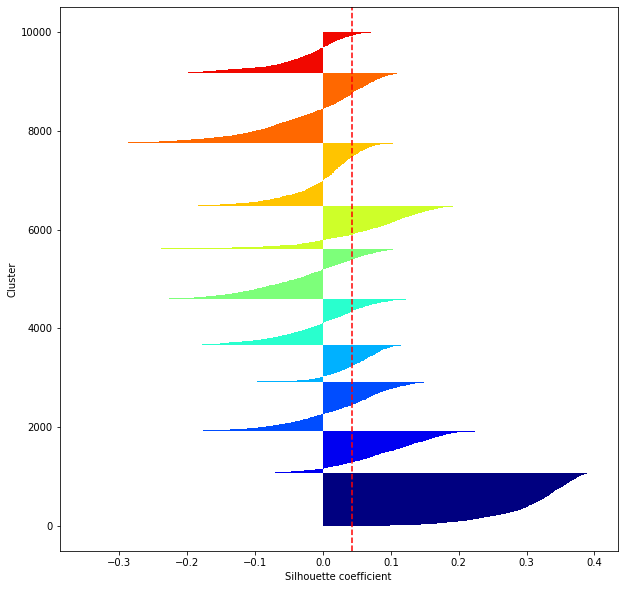
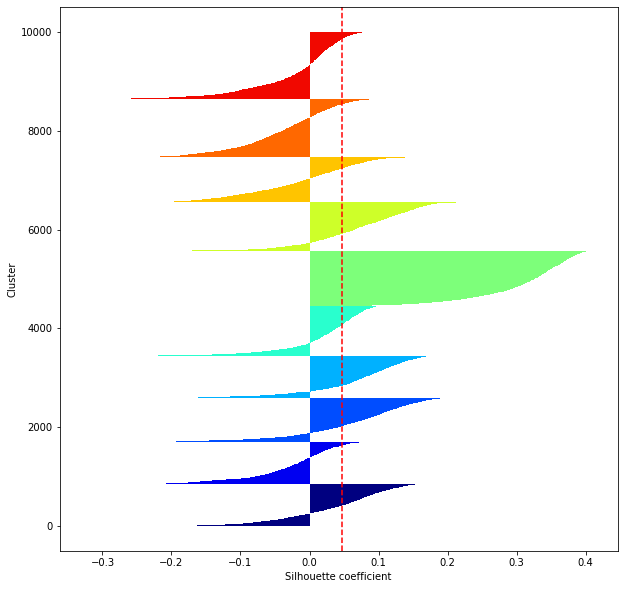
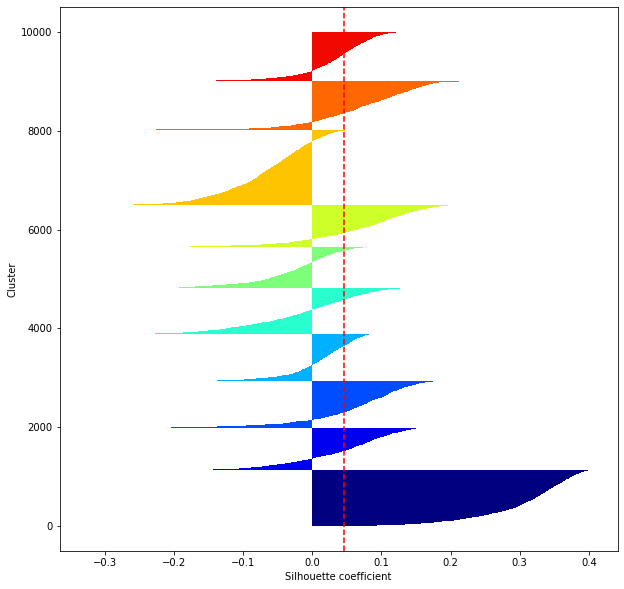
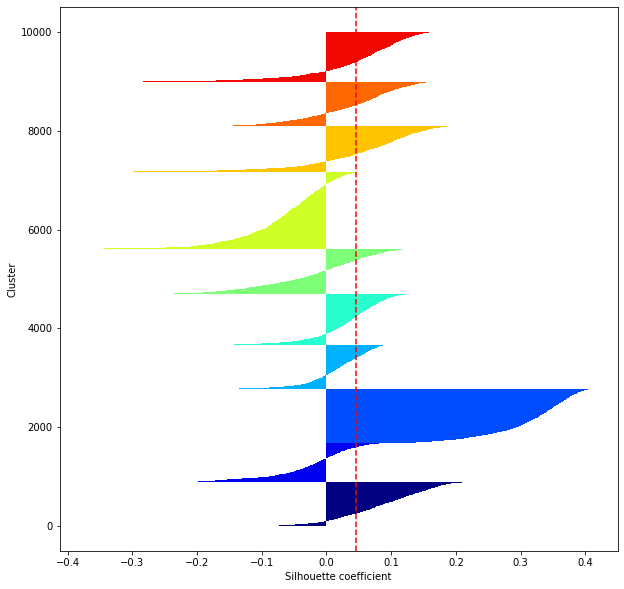
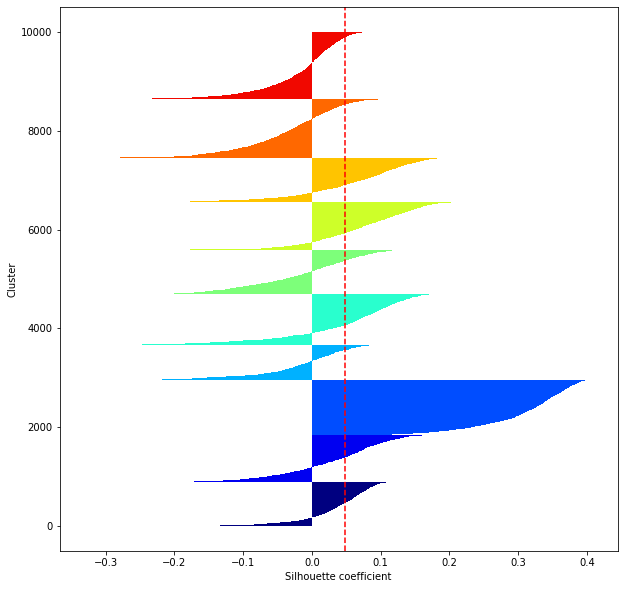
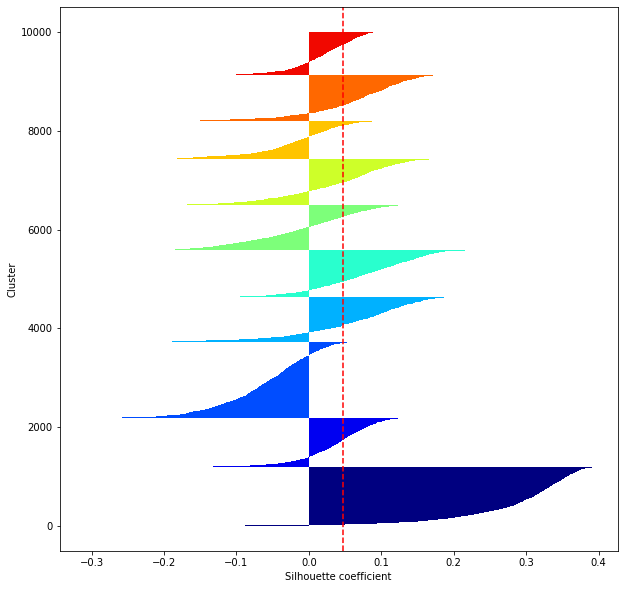
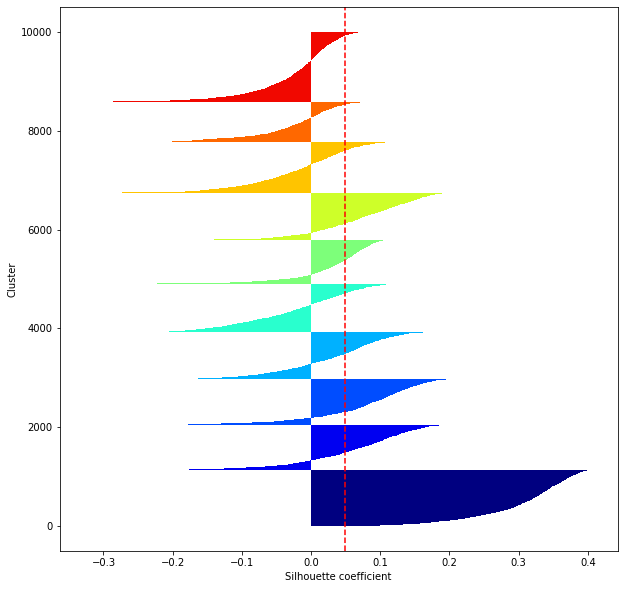
def shilhouette(clusters, x_test):
from sklearn.metrics import silhouette_samples
from matplotlib import cm
plt.figure(figsize = (10, 10))
cluster_labels=np.unique(clusters)
n_clusters=cluster_labels.shape[0]
silhouette_vals=silhouette_samples(x_test,clusters,metric='euclidean')
y_ax_lower,y_ax_upper=0,0
yticks=[]
for i,c in enumerate(cluster_labels):
c_silhouette_vals=silhouette_vals[clusters==c]
print(len(c_silhouette_vals))
c_silhouette_vals.sort()
y_ax_upper +=len(c_silhouette_vals)
color=cm.jet(float(i)/n_clusters)
plt.barh(range(y_ax_lower,y_ax_upper),
c_silhouette_vals,
height=1.0,
edgecolor='none',
color=color
)
yticks.append((y_ax_lower+y_ax_upper)/2.)
y_ax_lower += len(c_silhouette_vals)
#シルエット係数が1であれば 良くクラスタリングできてる
#またシルエットの幅がクラスタ数で平均して等しいとき、全体のデータを等分割できていることを示す
#この分割幅=シルエットバーの幅が等しくなり、かつ、シルエット係数が1に近づくようにkを最適化することが設定手法として考えられる.
#平均の位置に線を引く
silhouette_avg=np.mean(silhouette_vals)
plt.axvline(silhouette_avg,color="red",linestyle="--")
plt.ylabel("Cluster")
plt.xlabel("Silhouette coefficient")
for i in range(len(feature_dims)):
shilhouette(kms[i], x_test)
1228
2862
388
406
760
398
304
1653
1194
807
1350
1053
620
969
1312
1081
1720
544
270
1081
633
753
2140
711
320
786
926
1060
888
1783
1076
842
998
740
938
1022
861
1265
1425
833
855
850
884
858
1007
1118
979
911
1180
1358
1132
860
936
962
923
835
844
1533
974
1001
891
784
1096
892
1040
910
1562
914
904
1007
890
947
1127
710
1018
902
960
902
1178
1366
1189
1002
1538
903
947
918
925
771
927
880
1133
914
923
950
986
890
954
1023
816
1411