MobileNetの構造についてはこちらにまとめておきました。
概要
- 複雑な深層学習モデル構築はKerasやPyTorch等を利用した実装が早くて楽
- でもそれではつまらない
- 簡単なモデルであればアマチュアスクラッチでも動きそう
- なので最低限必要となるtrain_test_split, OneHotEncoder, MobileNet V1を実装
- 使用ライブラリはNumpy, Matplotlib, Pyprind(学習進捗バー表示用)のみ
- データはCIFAR-10を使用
洗練された深層学習ライブラリによるモデルの機能や構造とは異なる点が複数あります。
あくまで論文を元にしたスクラッチ学習・モデル理解を目的とした実装ですので、「モデルを使って何かしたい」という場合は素直に深層学習ライブラリを利用した方が無難です。
Python機械学習プログラミング、ゼロから作るDeepLearningのコードを適宜利用させていただいております。
対象読者
「本で読んだスクラッチ実装だけでは何か物足りない」というただ一点において共感していただける方々に有益な情報かと思います。
やはりスクラッチは面白いですね。
この調子でV2, V3も(時間が許す限り)実装していきたいです!
非対象読者
挙げきれないですが、ざっくり以下の通りでしょうか...。
- 深層学習ライブラリと併用したい
- Edge TPUで使いたい
- 転移学習したい
- include_top=Falseにして簡単に別の層群に付け替えたい
- 学習したモデル、重みを保存したい
注意点
- エラー処理の詳細な条件検討はしていません。
- ReLU6は未実装(単純な失念だけど、いずれにせよ量子化・軽量化まで考えると実装は難しそう)
- 重みの保存くらいは将来できそうだが未実装
- うまく動いているだけで、実装としては間違っている場合はご指摘ください。
評価結果
先に評価結果を出しておきます。
モデルとしてちゃんと動くかどうかをまずは見たかったので、とりあえず精度は度外視し、データサンプル数は全体のごく一部しか使用していません。
パラメータ調整も適当です。
データや実験方法に関してはこちらを参考にしています。
Google Colaboratoryでやってますが、メモリが少なすぎるのがきついですね。無料なので文句言えませんが...。
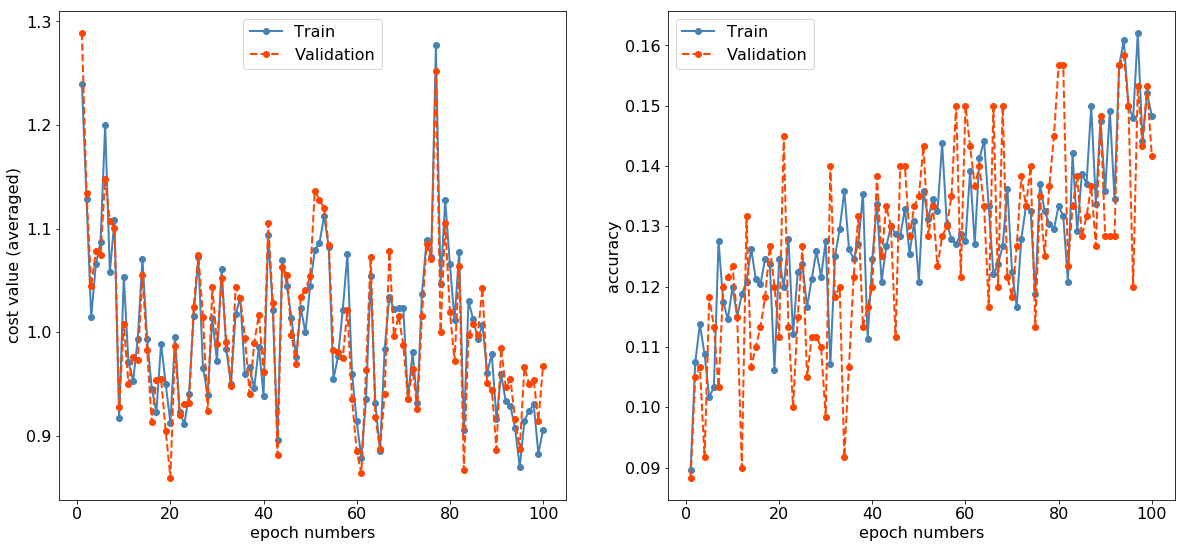
SGD evaluation (Learning rate: 0.01)
データ数
訓練データ数:2400
検証データ数:600
モデル構造
学習の推移
Adam evaluation (Learning rate: 0.001)
データ数
訓練データ数:2400
検証データ数:600
モデル構造
学習の推移
サンプル数が少なすぎるのでグラフが非常に粗いですが、コストが下がり、正解率が上がる傾向は一応見えているので、たぶんうまく実装できているものと思われます。
テストデータの推論時間は、平均で約5.8ms/imageでした。データ数が少ないのでなんとも言えませんが...。
今後は条件を振って、揃えて、スピードや精度を深層学習ライブラリと比較していきたいです。
コード
train_test_split
今回はあまり使っていませんが、scikit-learnのソースコードや実際の出力結果を比較して、だいたい同じ挙動となっていることは確認しています。
def train_test_split(X,y,test_size=0.25,
random_state=None,shuffle=True,stratify=None):
"""
Split the data to be learned and tested.
Parameters
----------
X : ndarray, shape (n_samples, n_features)
data to be learned and tested
y : ndarray, shape (n_samples, )
objective labels
test_size : float (0<test_size<1)(default: 0.25)
set the rate of test size
random_state : int
set the pseudo-random number to be used in RandomStateGenerator
shuffle : boolean (default:True)
shuffle before split or not. If False, set stratify as None.
stratify : array-like or None
array for stratified sampling
Returns
----------
X_train : ndarray, shape (n_samples, n_features)
data to be learned
X_test : ndarray, shape (n_samples, n_features)
data to be tested
y_train : ndarray, shape (n_samples, )
labels for X_train
y_test : ndarray, shape (n_samples, )
labels for X_test
"""
# Error if feature samples number does not corresponds to y number.
if X.shape[0] != y.shape[0]:
raise ValueError("X samples number({}) is not same as y {}.".format(
X.shape[0], y.shape[0]))
# make several parameters to be used
n_samples = X.shape[0]
n_train = np.floor((1-test_size) * n_samples).astype(int)
n_test = n_samples - n_train
classes = np.unique(y)
n_classes = len(classes)
class_counts = np.bincount(y)
class_indices = np.split(np.argsort(y, kind='mergesort'),
np.cumsum(class_counts)[:-1])
# Case1: Shuffle=False and stratify=None
if shuffle is False and stratify is None:
X_test = X[:n_test]
X_train = X[n_test:(n_test + n_train)]
y_test = y[:n_test]
y_train = y[n_test:(n_test + n_train)]
return X_train, X_test, y_train, y_test
# Case2: Shuffle=False and stratify=y
elif shuffle is False and stratify is not None:
raise ValueError("If 'shuffle' parameter is False, "
"then 'stratify' parameter should be None.")
# Case3: Shuffle=True and stratify=None
elif shuffle is True and stratify is None:
rng = np.random.RandomState(seed=random_state)
# shuffle and split
permutation = rng.permutation(n_samples)
ind_test = permutation[:n_test]
ind_train = permutation[n_test:(n_test + n_train)]
X_train = X[ind_train]
X_test = X[ind_test]
y_train = y[ind_train]
y_test = y[ind_test]
yield X_train
yield X_test
yield y_train
yield y_test
# Case4: Shuffle=True and stratify=y
else:
# set a number of samples to be selected per each class
rng = np.random.RandomState(seed=random_state)
n_i = _extracting_func(class_counts, n_train, rng)
class_counts_remaining = class_counts - n_i
t_i = _extracting_func(class_counts_remaining, n_test, rng)
train = []
test = []
# select at random which indices should be assigned to train and test set
for i in range(n_classes):
permutation = rng.permutation(class_counts[i])
perm_indices_class_i = class_indices[i].take(
permutation,mode='clip')
train.extend(perm_indices_class_i[:n_i[i]])
test.extend(perm_indices_class_i[n_i[i]:n_i[i] + t_i[i]])
ind_train = rng.permutation(train)
ind_test = rng.permutation(test)
X_train = X[ind_train]
X_test = X[ind_test]
y_train = y[ind_train]
y_test = y[ind_test]
yield X_train
yield X_test
yield y_train
yield y_test
def _extracting_func(class_counts, n_draws, rng):
"""
Internal function for "train_test_split" in "Case4: Shuffle=True and stratify=y"
Stratified sampling at random a certain number(n_draws) of samples
from population in class_counts.
"""
# assign each number of samples to be extracted per each class
continuous = n_draws * (class_counts / class_counts.sum())
floored = np.floor(continuous)
need_to_add = int(n_draws - floored.sum())
# determine which classes should be added one more because of flooring
if need_to_add > 0:
remainder = continuous - floored
# sort the remaining values in an unascending manner
values = np.sort(np.unique(remainder))[::-1]
for value in values:
inds, = np.where(remainder == value)
# set the number of value to be added
add_now = min(len(inds), need_to_add)
# determine at random where should be added
inds = rng.choice(inds, size=add_now, replace=False)
floored[inds] += 1
# repeat until when 'need to add' becomes 0
need_to_add -= add_now
if need_to_add == 0:
break
return floored.astype(np.int)
OneHotEncoder
これは直感のままシンプルに書いています。
class OneHotEncoder:
def fit(self, y):
self.max_value = np.max(y) + 1
def transform(self, y):
y_onehot = np.zeros((y.shape[0], self.max_value))
for i in range(y.shape[0]):
y_onehot[i, y[i]] += 1.
return y_onehot
def fit_transform(self, y):
self.fit(y)
y_onehot = self.transform(y)
return y_onehot
MobileNet V1
"MobileNet_v1"の中で"SeparableConvLayers", "ConvLayer", "FullConnLayer"(全結合層)などを呼んでいます。"SeparableConvLayers"の中ではさらに"ConvLayer", "BatchNormalization", "Activator"を呼んでいます。
List of classes and functions
- class MobileNet_v1
- class SeparableConvLayers
- class ConvLayer
- class BatchNormalization
- class PoolLayer
- def im2col
- def col2im
- class Flatten
- class Dropout
- class FullConnLayer
- class LearningMethod
- class SimpleInitializer
- class Activator
- class Softmax
- class GetMiniBatch
class MobileNet_v1:
"""
List of instances (Forward propagation ascending order)
----------
- ConvLayer
- DepthwiseSeparableConvLayers * 13
- PoolLayer
- Flatten
- Dropout
- FullConnLayer (without ReLU)
- Softmax
Parameters
----------
input_shape : tuple
Shape of input data(eg. (3, 224, 224))
alpha : float (0.25, 0.50, 0.75 or 1.0)
Width of the network (width multiplier) decreasing the number of filters in each layer.
dropout : float
dropout rate
include_top : int
Whether to include the fully-connected layer at the top of the network.
pooling : str
PoolLayer mode ("Max" or "Average")
n_classes : int
Number of classes for inference
learning_method : str
Optimization method ("SGD", "Momentum", "AdaGrad" or "Adam")
lr : float
Learning rate
sgd_momentum : float
Momentum parameter in "Momentum SGD"
adam_beta1 : float
Beta1 parameter in "Adam"
adam_beta2 : float
Beta2 parameter in "Adam"
batch_size : int
Mini-batch size
sigma : str or float
SD for initial value of weights of each layer ("Xavier", "He" or float value)
activation : str
Activation function ("sigmoid", "tanh" or "ReLU")
gamma : float
Scaling parameter for normalized data in batchnormalization
beta : float
Shifting parameter for normalized data in batchnormalization
momentum : float
Momentum for the moving mean and the moving variance in batchnormalization.
random_seed : int (default : None)
Pseudo random seed for data shuffling in each epoch
verbose : bool (default : True)
Whether to export learning proccess or not
Attributes
----------
self.train_cost_ : list
Cost value per epoch for train data(Cross entropy error)
self.val_cost_ : list
Cost value per epoch for validation data(Cross entropy error)
self.train_accuracy_ : list
Accuracy value per epoch for train data
self.val_accuracy_ : list
Accuracy value per epoch for validation data
"""
def __init__(self, input_shape=(3, 224, 224), alpha=1., dropout=1e-3,
pooling="Average", n_classes=1000,
learning_method="SGD", lr=0.01, sgd_momentum=0.9, adam_beta1=0.9, adam_beta2=0.999,
batch_size=10, sigma="Xavier", activation="ReLU",
gamma=1., beta=0., momentum=0.9,
n_epochs = 10, random_seed=0, verbose=True):
self.input_shape = input_shape
self.alpha = alpha
if alpha != 0.25 and alpha != 0.50 and alpha != 0.75 and alpha != 1.0:
raise ValueError("Set alpha to 0.25, 0.50, 0.75 or 1.0 .")
self.dropout = dropout
self.pooling = pooling
self.n_classes = n_classes
self.learning_method = learning_method
self.lr = lr
self.sgd_momentum = sgd_momentum
self.adam_beta1 = adam_beta1
self.adam_beta2 = adam_beta2
self.batch_size = batch_size
self.sigma = sigma
self.activation = activation
self.gamma = gamma
self.beta = beta
self.momentum = momentum
self.n_epochs = n_epochs
self.batch_size = batch_size
self.random_seed = random_seed # For mini-batch training
self.verbose = verbose
def build(self, show_model=True):
"""
Building model for classification
Parameters
----------
show_model : bool
Whether to show the summary of the model built
"""
# For cost and accuracy value
self.train_cost_ = []
self.val_cost_ = []
self.train_accuracy_ = []
self.val_accuracy_ = []
# Height of images, number of features(pixels) in an image
image_size = self.input_shape[1]
n_features = (image_size**2) * self.input_shape[0]
# Create an optimizer instance
optimizer = LearningMethod(self.learning_method, self.lr, self.sgd_momentum, self.adam_beta1, self.adam_beta2)
# Create instances of initial layers
self.ConvLayer = ConvLayer(
FN=int(32*self.alpha), C=3, FS=3, sigma=self.sigma, n=n_features, optimizer=optimizer, S=2, P=1)
self.BatchNormalization = BatchNormalization(gamma=self.gamma, beta=self.beta, momentum=self.momentum)
self.Activator = Activator(self.activation)
# Prepare parameters for a series of SeparableConvLayers
FN_list = [32,64,64,128,128,128,128,256,256,256,256,512,512,512,512,512,512,512,512,512,512,512,512,1024,1024,1024]
C_list = [32,32,64,64,128,128,128,128,256,256,256,256,512,512,512,512,512,512,512,512,512,512,512,512,1024,1024]
FS_list = [3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1,3,1]
input_size_list = [112,112,112,56,56,56,56,28,28,28,28,14,14,14,14,14,14,14,14,14,14,14,14,7,7,7]
S_list = [1,1,2,1,1,1,2,1,1,1,2,1,1,1,1,1,1,1,1,1,1,1,2,1,2,1]
n_list = []
# Adjust parameters with alpha
for index in np.arange(26):
FN_list[index] = int(FN_list[index] * self.alpha)
C_list[index] = int(C_list[index] * self.alpha)
input_size_list[index] = int(input_size_list[index] * (image_size/224))
n_list.append(((input_size_list[index])**2) * C_list[index])
# Create instances of a series of SeparableConvLayers
self.SCLayers = []
for index in np.arange(0,26,2):
self.SCLayers.append(SeparableConvLayers(
optimizer, self.sigma, self.activation,
FN_list[index], C_list[index], FS_list[index], n_list[index], S_list[index], 1,
FN_list[index+1], C_list[index+1], FS_list[index+1], n_list[index+1], S_list[index+1], 0,
self.gamma, self.beta, self.momentum))
# Create instances of final layers
self.PoolLayer = PoolLayer(PH=input_size_list[-1], PW=input_size_list[-1], pooling=self.pooling)
self.Flatten = Flatten()
self.Dropout = Dropout(prob=self.dropout)
self.FullConnLayer = FullConnLayer(FN_list[-1], self.n_classes,
SimpleInitializer(self.sigma, FN_list[-1]), optimizer)
self.Softmax = Softmax()
# If model should be visualized,
if show_model:
Layer_names = ["Conv / s2 ",
"Conv dw / s1","Conv / s1 ","Conv dw / s2","Conv / s1 ",
"Conv dw / s1","Conv / s1 ","Conv dw / s2","Conv / s1 ",
"Conv dw / s1","Conv / s1 ","Conv dw / s2","Conv / s1 ",
"Conv dw / s1","Conv / s1 ","Conv dw / s1","Conv / s1 ",
"Conv dw / s1","Conv / s1 ","Conv dw / s1","Conv / s1 ",
"Conv dw / s1","Conv / s1 ",
"Conv dw / s2","Conv / s1 ","Conv dw / s2","Conv / s1 ",
"{} Pool / s1".format(self.pooling),"FC / s1 ","Softmax / s1"
]
Input_channels = [self.input_shape[0], ] + C_list + [C_list[-1], C_list[-1], self.n_classes]
Input_sizes = [self.input_shape[1], ] + input_size_list + [input_size_list[-1],1,1]
Output_channels = [int(32*self.alpha), ] + FN_list + [FN_list[-1], self.n_classes, self.n_classes]
Output_sizes = input_size_list + [input_size_list[-1],1,1,1]
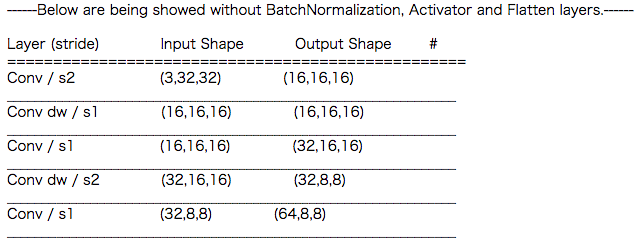
print("------Below are being showed without BatchNormalization, Activator and Flatten layers.------\n")
print("Layer (stride) Input Shape Output Shape # ")
print("==================================================")
for i in range(30):
print("{} ({},{},{}) ({},{},{})".format(
Layer_names[i],
Input_channels[i],
Input_sizes[i],
Input_sizes[i],
Output_channels[i],
Output_sizes[i],
Output_sizes[i]))
if i != 29:
print("________________________________________________________________")
print("==================================================")
print("shape: (C, H, W)")
def fit(self, X_train, y_train, X_val=None, y_val=None):
"""
Fitting (training) for classification
Parameters
----------
X_train : ndarray, shape (n_samples, channels, image_height, image_width)
Features of train data
y_train : ndarray, shape (n_samples, )
Labels of train data
X_val : ndarray, shape (n_samples, channels, image_height, image_width)
Features of validation data
y_val : ndarray, shape (n_samples, )
Labels of validation data
"""
# Labels OneHotEncoding
enc = OneHotEncoder()
y_train_onehot = enc.fit_transform(y_train[:, np.newaxis])
y_val_onehot = enc.transform(y_val[:, np.newaxis])
# Set progress bar for training
pbar = pyprind.ProgBar(self.n_epochs * (np.ceil(X_train.shape[0] / self.batch_size).astype(np.int)))
# Create a random_seeds array
rgn = np.random.RandomState(self.random_seed)
random_seeds = np.arange(self.n_epochs)
rgn.shuffle(random_seeds)
# Train per epoch
for i in range(self.n_epochs):
get_mini_batch = GetMiniBatch(X_train, y_train_onehot,
batch_size=self.batch_size, seed=random_seeds[i])
# Create batch data
for X_train_batch, y_train_batch in get_mini_batch:
# Forward propagation
z_out, a_out = self._forward(X_train_batch, train_flg=True)
# Backward propagation
dx = self.Softmax.backward(a_out, y_train_batch) # including cross entropy error and softmax
dx = self.FullConnLayer.backward(dx)
dx = self.Dropout.backward(dx)
dx = self.Flatten.backward(dx)
dx = self.PoolLayer.backward(dx)
for i in np.arange(12, -1, -1):
dx = self.SCLayers[i].backward(dx)
dx = self.Activator.backward(dx)
dx = self.BatchNormalization.backward(dx)
dx = self.ConvLayer.backward(dx)
# Update progress bar
pbar.update()
# Save cost and accuracy of train data per epoch
z_out, a_out = self._forward(X_train, train_flg=False)
# Cost value
value1 = y_train_onehot * (np.log(a_out + 1e-07)) # for ZeroDivision error
value2 = (1. - y_train_onehot) * np.log(1. - a_out + 1e-07) # for ZeroDivision error
train_cost = -np.sum(value1 + value2)
self.train_cost_.append(train_cost)
# Accuracy
y_train_pred = self.predict(X_train)
train_accuracy = (np.sum(y_train == y_train_pred).astype(np.float)) / y_train.shape[0]
self.train_accuracy_.append(train_accuracy)
# Save as well if there are validation data
if X_val is not None and y_val is not None:
z_out, a_out = self._forward(X_val, train_flg=False)
# Cost value
value1 = y_val_onehot * (np.log(a_out + 1e-07)) # for ZeroDivision error
value2 = (1. - y_val_onehot) * np.log(1. - a_out + 1e-07) # for ZeroDivision error
val_cost = -np.sum(value1 + value2)
self.val_cost_.append(val_cost)
# Accuracy
y_val_pred = self.predict(X_val)
val_accuracy = (np.sum(y_val == y_val_pred).astype(np.float)) / y_val.shape[0]
self.val_accuracy_.append(val_accuracy)
# Each cost value should be divided by mean value
self.train_cost_ /= (sum(self.train_cost_) / len(self.train_cost_))
if len(self.val_cost_) > 0:
self.val_cost_ /= (sum(self.val_cost_) / len(self.val_cost_))
# If verbose, export the train proccess
if self.verbose:
plt.rcParams["font.size"] = 16
fig, ax = plt.subplots(1, 2, figsize=(20,9))
ax[0].plot(np.arange(1, len(self.train_cost_)+1), self.train_cost_,
linewidth=2, linestyle="-", marker="o", color="steelblue", label="Train")
ax[0].set_xlabel("epoch numbers")
ax[0].set_ylabel("cost value (averaged)")
ax[1].plot(np.arange(1, len(self.train_accuracy_)+1), self.train_accuracy_,
linewidth=2, linestyle="-", marker="o", color="steelblue", label="Train")
ax[1].set_xlabel("epoch numbers")
ax[1].set_ylabel("accuracy")
# Export as well if there are validation data
if len(self.val_cost_) > 0:
ax[0].plot(np.arange(1, len(self.val_cost_)+1), self.val_cost_,
linewidth=2, linestyle="--", marker="o", color="orangered", label="Validation")
ax[1].plot(np.arange(1, len(self.val_accuracy_)+1), self.val_accuracy_,
linewidth=2, linestyle="--", marker="o", color="orangered", label="Validation")
ax[0].legend()
ax[1].legend()
plt.show()
def predict(self, X_test):
z_out, a_out = self._forward(X_test, train_flg=False)
return np.argmax(z_out, axis=1) # Export max-value index in input data for Softmax layer
def _forward(self, x, train_flg):
# Forward propagation
x = self.ConvLayer.forward(x)
x = self.BatchNormalization.forward(x, train_flg)
x = self.Activator.forward(x)
for i in np.arange(13):
x = self.SCLayers[i].forward(x, train_flg)
x = self.PoolLayer.forward(x)
x = self.Flatten.forward(x)
x = self.Dropout.forward(x)
z_out = self.FullConnLayer.forward(x)
a_out = self.Softmax.forward(z_out)
return z_out, a_out
class SeparableConvLayers:
"""
List of instances (Forward propagation ascending order)
----------
- ConvLayer (Depthwise)
- BatchNormalization
- Activator (ReLU)
- ConvLayer (Pointwise)
- BatchNormalization
- Activator (ReLU)
Parameters
----------
optimizer : an instance
An instance of a class "LearningMethod"
sigma : str or float
SD for initial value of weights of each layer ("Xavier", "He" or float value)
activation : str
Activation function ("sigmoid", "tanh" or "ReLU")
train_flg : bool
Whether Batchnormalization layer is in train phase or not
gamma : float
Scaling parameter for normalized data
beta : float
Shifting parameter for normalized data
momentum : float
Momentum for the moving mean and the moving variance in BatchNormalization.
"""
def __init__(self, optimizer, sigma, activation,
FN_depth, C_depth, FS_depth, n_depth, S_depth, P_depth,
FN_point, C_point, FS_point, n_point, S_point, P_point,
gamma, beta, momentum):
# Set each layers
# Depthwise convolution
self.DepthwiseConvLayer = ConvLayer(
FN_depth, C_depth, FS_depth, sigma, n_depth, optimizer, S=S_depth, P=P_depth, depthwise=True)
self.BatchNormalization_depth = BatchNormalization(
gamma=gamma, beta=beta, momentum=momentum)
self.Activator_depth = Activator(activation)
# Pointwise convolution
self.PointwiseConvLayer = ConvLayer(
FN_point, C_point, FS_point, sigma, n_point, optimizer, S=S_point, P=P_point)
self.BatchNormalization_point = BatchNormalization(
gamma=gamma, beta=beta, momentum=momentum)
self.Activator_point = Activator(activation)
def forward(self, x, train_flg):
x = self.DepthwiseConvLayer.forward(x)
x = self.BatchNormalization_depth.forward(x, train_flg)
x = self.Activator_depth.forward(x)
x = self.PointwiseConvLayer.forward(x)
x = self.BatchNormalization_point.forward(x, train_flg)
x = self.Activator_point.forward(x)
return x
def backward(self, dx):
dx = self.Activator_point.backward(dx)
dx = self.BatchNormalization_point.backward(dx)
dx = self.PointwiseConvLayer.backward(dx)
dx = self.Activator_depth.backward(dx)
dx = self.BatchNormalization_depth.backward(dx)
dx = self.DepthwiseConvLayer.backward(dx)
return dx
class ConvLayer:
"""
Convolution layer class
Parameters
----------
FN : int
Output channels (filter number)
C : int
Input channels
FS : int
Filter size (number of elements(pixels))
sigma : str, float or int
SD for initial value of weights ("Xavier", "He" or float value)
n : int
Total node number in previous layer or number of features (input image)
optimizer : instance
An instance of a class "LearningMethod"
S : int (default: 1)
Stride
P : int (default: 0)
Padding
depthwise : bool
Whether depthwise convolution or not
Attributes
----------
self.z_shape : input dimensions
Input data dimensions preserved for backward propagation
"""
def __init__(self, FN, C, FS, sigma, n, optimizer, S=1, P=0, depthwise=False):
self.S = S
self.P = P
self.optimizer = optimizer
self.depthwise = depthwise
# Set sigma to float value
if (type(sigma) is float) or (type(sigma) is int):
self.sigma = float(sigma)
elif sigma == "Xavier":
self.sigma = np.sqrt(1. / n)
elif sigma == "He":
self.sigma = np.sqrt(2. / n)
else:
raise ValueError("Set \"Xavier\" or \"He\" or numerical value (float or int).")
# self.w and self.b initialization
self.w = self.sigma * np.random.randn(FN, C, FS, FS)
# Mask weights when depthwise convolution
if self.depthwise:
self.mask = np.zeros((FN, C, FS, FS))
for mask in range(FN):
# FN = C when depthwise conv
self.mask[mask, mask, :, :] += 1
self.w *= self.mask
self.b = np.zeros(FN)
# Create parameter "v" when Momentum optimization
if self.optimizer.learning_method == "Momentum":
self.v_w = np.zeros((FN, C, FS, FS))
self.v_b = np.zeros(FN)
# Create parameter "h" for learning coefficients decay when AdaGrad optimization
elif self.optimizer.learning_method == "AdaGrad":
self.h_w = np.zeros((FN, C, FS, FS))
self.h_b = np.zeros(FN)
# Create parameter "m" and "v" when Adam optimization
elif self.optimizer.learning_method == "Adam":
self.m_w = np.zeros((FN, C, FS, FS))
self.v_w = np.zeros((FN, C, FS, FS))
self.m_b = np.zeros(FN)
self.v_b = np.zeros(FN)
def forward(self, z):
FN, C, FH, FW = self.w.shape # (Output channels, Input channels, Filter height, filter width)
N, C, H, W = z.shape # (Number of batch data, Input channels, Input image height, Input image width)
OH = self._conv_func(H, self.P, FH, self.S) # Output height
OW = self._conv_func(W, self.P, FW, self.S) # Output width
col = im2col(z, FH, FW, self.S, self.P)
col_w = self.w.reshape(FN, -1).T
# Inner product (N * OH * OW, C * FH * FW) and (C * FS * FS, FN) to (N * OH * OW, FN)
a = np.dot(col, col_w) + self.b
# Split N * OH * OW into each and transpose to "NCHW" standard shape
a = a.reshape(N, OH, OW, -1).transpose(0, 3, 1, 2)
# Save for backward propagation
self.z_shape = z.shape
self.col = col # N, OH, OW in axis 0, C, FH, FW in axis 1
self.col_w = col_w # shape (C * FS * FS, FN)
return a
def backward(self, grad_a):
FN, C, FH, FW = self.w.shape # (Output channels, Input channels, Filter height, filter width)
N, C, H, W = self.z_shape # (Number of batch data, Input channels, Input image height, Input image width)
# Traspose to (N, OH, OW, FN) and reshape to (N * OH * OW, FN)
grad_a = grad_a.transpose(0,2,3,1).reshape(-1, FN)
# b gradient calculation
self.grad_b = np.sum(grad_a, axis=0)
# w gradient calculation
# Inner product (C * FH * FW, N * OH * OW) and (N * OH * OW, FN) to (C * FH * FW, FN)
self.grad_w = np.dot(self.col.T, grad_a)
# Transpose to (FN, C * FH * FW) and reshape to (FN, C, FH, FW) standard shape
self.grad_w = self.grad_w.transpose(1, 0).reshape(FN, C, FH, FW)
# Mask gradients when depthwise convolution
if self.depthwise:
self.grad_w *= self.mask
# z gradient calculation
# Inner product (N * OH * OW, FN) and (FN, C * FS * FS) to (N * OH * OW, C * FS * FS)
grad_col = np.dot(grad_a, self.col_w.T)
# Adjust to "NCHW" standard shape with col2im
grad_z = col2im(grad_col, self.z_shape, FH, FW, self.S, self.P)
# weights update
self = self.optimizer.update(self)
return grad_z
def _conv_func(self, N, P, F, S):
# Output size calculation with input image size (N), padding (P), filter size (F), stride (S)
return int(((N + 2 * P - F)/ S) + 1)
class BatchNormalization:
"""
http://arxiv.org/abs/1502.03167
"""
def __init__(self, gamma, beta, momentum, running_mean=None, running_var=None):
self.gamma = gamma
self.beta = beta
self.momentum = momentum
self.input_shape = None # 4 dimensions in Conv layer and 2 dimensions in Dense layer
# mean and variance to be used in test phase
self.running_mean = running_mean
self.running_var = running_var
# for backward propagation
self.batch_size = None
self.xc = None
self.std = None
self.dgamma = None
self.dbeta = None
def forward(self, x, train_flg):
self.input_shape = x.shape
if x.ndim != 2:
N, C, H, W = x.shape
x = x.reshape(N, -1)
out = self.__forward(x, train_flg)
return out.reshape(*self.input_shape)
def __forward(self, x, train_flg):
if self.running_mean is None:
N, D = x.shape
self.running_mean = np.zeros(D)
self.running_var = np.zeros(D)
if train_flg:
mu = x.mean(axis=0)
xc = x - mu
var = np.mean(xc**2, axis=0)
std = np.sqrt(var + 10e-7)
xn = xc / std
self.batch_size = x.shape[0]
self.xc = xc
self.xn = xn
self.std = std
self.running_mean = self.momentum * self.running_mean + (1-self.momentum) * mu
self.running_var = self.momentum * self.running_var + (1-self.momentum) * var
else:
xc = x - self.running_mean
xn = xc / ((np.sqrt(self.running_var + 10e-7)))
out = self.gamma * xn + self.beta
return out
def backward(self, dout):
if dout.ndim != 2:
N, C, H, W = dout.shape
dout = dout.reshape(N, -1)
dx = self.__backward(dout)
dx = dx.reshape(*self.input_shape)
return dx
def __backward(self, dout):
dbeta = dout.sum(axis=0)
dgamma = np.sum(self.xn * dout, axis=0)
dxn = self.gamma * dout
dxc = dxn / self.std
dstd = -np.sum((dxn * self.xc) / (self.std * self.std), axis=0)
dvar = 0.5 * dstd / self.std
dxc += (2.0 / self.batch_size) * self.xc * dvar
dmu = np.sum(dxc, axis=0)
dx = dxc - dmu / self.batch_size
self.dgamma = dgamma
self.dbeta = dbeta
return dx
class PoolLayer:
"""
Parameters
----------
PH(Pooling Height) : int
Height of pooling kernels
PW(Pooling Width) : int
Width of pooling kernels
PS(Pooling Stride) : int
Stride value of pooling kernels
PP(Pooling Padding) : int
Padding value of pooling kernels
pooling : str
"Max" or "Average"
Attributes
----------
self.z_shape : tuple of ints
Dimension values of input data saved for calculation of backward propagation
"""
def __init__(self, PH, PW, PS=1, PP=0, pooling="Average"):
self.PH = PH
self.PW = PW
self.PS = PS
self.PP = PP
self.pooling = pooling
if self.pooling != "Max" and self.pooling != "Average":
raise ValueError("Set \"Max\" or \"Average\".")
def forward(self, z):
N, C, H, W = z.shape
OH = int(1 + (H - self.PH) / self.PS)
OW = int(1 + (W - self.PW) / self.PS)
# (N, C, H, W) to (N * OH * OW, C * FH * FW)
# At this moment, arrays correspond to each pooling-applied area are already alligned along axis 1.
col = im2col(z, self.PH, self.PW, self.PS, self.PP)
# Set size of axis 1 to that of each pooling-applied area
col = col.reshape(-1, self.PH * self.PW)
if self.pooling == "Max":
a = np.max(col, axis=1)
a = a.reshape(N, OH, OW, C).transpose(0, 3, 1, 2)
# Save for backward propagation
self.z_shape = z.shape
self.arg_max = np.argmax(col, axis=1) # Index of max value to be learned for backward propagation
elif self.pooling == "Average":
a = np.mean(col, axis=1)
a = a.reshape(N, OH, OW, C).transpose(0, 3, 1, 2)
# Save for backward propagation
self.z_shape = z.shape
self.col_shape = col.shape[0]
return a
def backward(self, grad_a):
# Reshape to (N, OH, OW, C)
grad_a = grad_a.transpose(0, 2, 3, 1)
pool_size = self.PH * self.PW
# Make a zero array with a size(Number of gradient elements, size of elements of pooling applied area)
grad_col = np.zeros((grad_a.size, pool_size))
if self.pooling == "Max":
# Update gradients only where values have been exploited when forward propagation
grad_col[np.arange(self.arg_max.size), self.arg_max.flatten()] = grad_a.flatten()
elif self.pooling == "Average":
# Update with mean values of gradients
grad_col += grad_a.flatten().reshape(-1,1)/pool_size
# Reshape to (N * OH * OW, C * FH * FW)
grad_col = grad_col.reshape(grad_a.shape + (pool_size,))
grad_col = grad_col.reshape(grad_col.shape[0] * grad_col.shape[1] * grad_col.shape[2], -1)
# Adjust to "NCHW" shape by col2im
grad_z = col2im(grad_col, self.z_shape, self.PH, self.PW, self.PS, self.PP)
return grad_z
def im2col(X, FH, FW, S=1, P=0):
"""
Parameters
----------
X : shape (n_samples, channels, heights, width)
FH : Filter height
FW : Filter width
S : Stride
P : Padding
Returns
-------
col : 2 dimensions ndarray
"""
N, C, H, W = X.shape
# Calculate pseudo output image height and width (4 dimensions)
OH = (H + 2 * P - FH)//S + 1
OW = (W + 2 * P - FW)//S + 1
img = np.pad(X, [(0,0), (0,0), (P, P), (P, P)], 'constant') # 高さと幅方向にパディング
col = np.zeros((N, C, FH, FW, OH, OW))
# Extracting every image data corresponds to each filter elements
for y in range(FH):
y_max = y + S * OH
for x in range(FW):
x_max = x + S * OW
col[:, :, y, x, :, :] = img[:, :, y:y_max:S, x:x_max:S]
# N, OH, OW in axis 0, C, FH, FW in axis 1
col = col.transpose(0, 4, 5, 1, 2, 3).reshape(N * OH * OW, -1)
return col
def col2im(col, X_shape, FH, FW, S=1, P=0):
"""
Parameters
----------
col : shape (N * OH * OW, C * FS * FS)
X_shape : Input data shape (e.g. (10, 1, 28, 28))
FH : Filter height
FW : Filter width
S : Stride
P : Padding
Returns
-------
X : shape(n_samples, channels, heights, width)
"""
# Inverse process compared to im2col
N, C, H, W = X_shape
OH = (H + 2 * P - FH)//S + 1
OW = (W + 2 * P - FW)//S + 1
col = col.reshape(N, OH, OW, C, FH, FW).transpose(0, 3, 4, 5, 1, 2)
img = np.zeros((N, C, H + 2 * P + S - 1, W + 2 * P + S - 1))
for y in range(FH):
y_max = y + S * OH
for x in range(FW):
x_max = x + S * OW
img[:, :, y:y_max:S, x:x_max:S] += col[:, :, y, x, :, :]
# Return "NCHW" standard shape (excluding padding edge)
X = img[:, :, P:H + P, P:W + P]
return X
class Flatten:
"""
Flatten class
Reduce dimensions (flattening) and make up data suitable for Dense layer inputs
Attributes
----------
self.z_shape : Input data dimensions
Dimension values of input data saved for calculation of backward propagation
"""
def forward(self, z):
# Vanish the dimensions of output channels and image size
self.z_shape = z.shape # for backward propagation
z = z.reshape(z.shape[0], z.shape[1] * z.shape[2] * z.shape[3])
return z
def backward(self, grad_a):
# Reshape for the previous layer
grad_z = grad_a.reshape(self.z_shape)
return grad_z
class Dropout():
def __init__(self, prob=1e-3):
self.prob = prob
def forward(self, X):
# Switch on and off using mask matrix
self.mask = np.random.binomial(1, 1. - self.prob, size=X.shape) / (1. - self.prob)
out = X * self.mask
return out
def backward(self, dout):
# Switch on and off using mask matrix
dX = dout * self.mask
return dX
class FullConnLayer:
"""
Dense layer class
Parameters
----------
n_nodes1 : int
Previous nodes total number
n_nodes2 : int
Next nodes total number
initializer : instance
An instance for initialization method
optimizer : instance
An instance for optimization method
Attributes
----------
self.z : shape (batch_size, n_nodes1)
Input data preserved for backward propagation
"""
def __init__(self, n_nodes1, n_nodes2, initializer, optimizer):
# self.w and self.b initialization using initializer
self.w = initializer.W(n_nodes1, n_nodes2)
self.b = initializer.B(n_nodes2)
self.optimizer = optimizer
# Create parameter "v" when Momentum optimization
if self.optimizer.learning_method == "Momentum":
self.v_w = np.zeros((n_nodes1, n_nodes2))
self.v_b = np.zeros(n_nodes2)
# Create parameter "h" for learning coefficients decay when AdaGrad optimization
elif self.optimizer.learning_method == "AdaGrad":
self.h_w = np.zeros((n_nodes1, n_nodes2))
self.h_b = np.zeros(n_nodes2)
# Create parameter "m" and "v" when Adam optimization
elif self.optimizer.learning_method == "Adam":
self.m_w = np.zeros((n_nodes1, n_nodes2))
self.v_w = np.zeros((n_nodes1, n_nodes2))
self.m_b = np.zeros(n_nodes2)
self.v_b = np.zeros(n_nodes2)
def forward(self, z):
"""
Parameters
----------
z : shape (batch_size, n_nodes1)
Input data
Returns
----------
a : shape (batch_size, n_nodes2)
Output data
"""
# Save input data and the dimensions for backward propagation
self.z = z
self.z_shape = z.shape
a = np.dot(z, self.w) + self.b
return a
def backward(self, grad_a):
"""
Parameters
----------
grad_a : shape (batch_size, n_nodes2)
Gradients flowed from behind
Returns
----------
grad_z : shape (batch_size, n_nodes1)
Gradients to be sent to the previous layer
"""
self.grad_w = np.dot(self.z.T, grad_a)
self.grad_b = grad_a.sum(axis=0)
grad_z = np.dot(grad_a, self.w.T)
# Update weights value
self = self.optimizer.update(self)
return grad_z
class LearningMethod:
"""
For an instance to be handled in to "Layer" class constructor as an optimization method.
Update layer's weights with "layer.----".
Parameters
----------
learning_method : str
"SGD", "Momentum", "AdaGrad" or "Adam"
lr : float
Learning rate
sgd_momentum : float
Momentum parameter in "Momentum SGD"
adam_beta1 : float
Beta1 parameter in "Adam"
adam_beta2 : float
Beta2 parameter in "Adam"
"""
def __init__(self, learning_method, lr, sgd_momentum=0.9, adam_beta1=0.9, adam_beta2=0.999):
self.learning_method = learning_method
self.lr = lr
self.sgd_momentum = sgd_momentum
self.adam_beta1 = adam_beta1
self.adam_beta2 = adam_beta2
self.iter = 0 # for "Adam" optimization
def update(self, layer):
if self.learning_method == "SGD":
layer.w -= self.lr * layer.grad_w / layer.z_shape[0]
layer.b -= self.lr * layer.grad_b / layer.z_shape[0]
return layer
elif self.learning_method == "Momentum":
layer.v_w = self.sgd_momentum * layer.v_w - self.lr * layer.grad_w / layer.z_shape[0]
layer.v_b = self.sgd_momentum * layer.v_b - self.lr * layer.grad_b / layer.z_shape[0]
layer.w += layer.v_w
layer.b += layer.v_b
return layer
elif self.learning_method == "AdaGrad":
layer.h_w += layer.grad_w * layer.grad_w
layer.h_b += layer.grad_b * layer.grad_b
layer.w -= self.lr * (1. / (np.sqrt(layer.h_w) + 1e-07)) * layer.grad_w / layer.z_shape[0]
layer.b -= self.lr * (1. / (np.sqrt(layer.h_b) + 1e-07)) * layer.grad_b / layer.z_shape[0]
return layer
elif self.learning_method == "Adam":
self.iter += 1
lr_t = self.lr * np.sqrt(1.0 - self.adam_beta2**self.iter) / (1.0 - self.adam_beta1**self.iter)
layer.m_w += (1 - self.adam_beta1) * (layer.grad_w - layer.m_w)
layer.v_w += (1 - self.adam_beta2) * (layer.grad_w**2 - layer.v_w)
layer.m_b += (1 - self.adam_beta1) * (layer.grad_b - layer.m_b)
layer.v_b += (1 - self.adam_beta2) * (layer.grad_b**2 - layer.v_b)
layer.w -= lr_t * layer.m_w / (np.sqrt(layer.v_w) + 1e-7) / layer.z_shape[0]
layer.b -= lr_t * layer.m_b / (np.sqrt(layer.v_b) + 1e-7) / layer.z_shape[0]
return layer
else:
raise ValueError("Set \"SGD\", \"Momentum\", \"AdaGrad\" or \"Adam\".")
class SimpleInitializer:
"""
An instance to be handled in to Dense layer (FullConnLayer class) constructor as initializing method.
Initializing using "self.W" and "self.B".
Parameters
----------
sigma : str, float or int
SD for initial value of weights of each layer ("Xavier", "He" or float value)
n : int
Nodes number in previous layer
"""
def __init__(self, sigma, n):
if (type(sigma) is float) or (type(sigma) is int):
self.sigma = float(sigma)
elif sigma == "Xavier":
self.sigma = np.sqrt(1. / n)
elif sigma == "He":
self.sigma = np.sqrt(2. / n)
else:
raise ValueError("Set \"Xavier\" or \"He\" or numerical value (float or int).")
def W(self, n_nodes1, n_nodes2):
return self.sigma * np.random.randn(n_nodes1, n_nodes2)
def B(self, n_nodes2):
return self.sigma * np.random.randn(n_nodes2)
class Activator:
"""
Activation function class
Parameters
----------
activation : str
Types of activation function ("sigmoid", "tanh" or "ReLU")
Attributes
----------
self.a : shape (batch_size, n_nodes2)
Input data preserved for backward propagation
"""
def __init__(self, activation):
self.activation = activation
def forward(self, a):
# Save input data "a" for backward propagation
self.a = a
if self.activation == "sigmoid":
sigmoid_range = 34.538776394910684
a = np.clip(a, -sigmoid_range, sigmoid_range) # for overflow
return 1 / (1 + np.exp(-a))
elif self.activation == "tanh":
return np.tanh(a)
elif self.activation == "ReLU":
return np.maximum(0, a)
else:
raise ValueError("Set \"sigmoid\" or \"tanh\" or \"ReLU\".")
def backward(self, grad_a):
if self.activation == "sigmoid":
return grad_a * (self.a * (1. - self.a))
elif self.activation == "tanh":
return grad_a * (1. - (np.tanh(self.a))**2)
elif self.activation == "ReLU":
derivative = np.where(self.a > 0., 1., 0.)
return grad_a * derivative
else:
raise ValueError("Set \"sigmoid\" or \"tanh\" or \"ReLU\".")
class Softmax:
"""
Softmax function class
"""
def forward(self, a):
a_max = np.max(a, axis=1)
exp_a = np.exp(a - a_max.reshape(-1, 1)) # a_max for overflow
sum_exp_a = np.sum(exp_a, axis=1).reshape(-1, 1)
return exp_a / sum_exp_a
def backward(self, a_out, y):
return a_out - y
class GetMiniBatch:
"""
Iterator to acquire mini-batch data
Acquire mini-batch data using "for" sentence in neural network
Parameters
----------
X : shape (n_samples, channels, image_height, image_width)
Train data
y : shape (n_samples, 1)
Labels
batch_size : int
Batch size
seed : int
Seed value for pseudo randomization
"""
def __init__(self, X, y, batch_size=10, seed=0):
self.batch_size = batch_size
np.random.seed(seed)
shuffle_index = np.random.permutation(np.arange(X.shape[0]))
self.X = X[shuffle_index]
self.y = y[shuffle_index]
self._stop = np.ceil(X.shape[0]/self.batch_size).astype(np.int)
# Return iteration times in 1 epoch
def __len__(self):
return self._stop
# Return batch data per iteration
def __getitem__(self, item):
p0 = item * self.batch_size
p1 = item * self.batch_size + self.batch_size
return self.X[p0:p1], self.y[p0:p1]
# Set iteration count value to zero
def __iter__(self):
self._counter = 0
return self
# Return batch data for next iteration
def __next__(self):
if self._counter >= self._stop:
raise StopIteration()
p0 = self._counter * self.batch_size
p1 = self._counter * self.batch_size + self.batch_size
self._counter += 1
return self.X[p0:p1], self.y[p0:p1]