蛍光X線プロファイル
最近は,蛍光X線の中心エネルギー,強度比,タイプ,自然幅は,xraylibを使えば,簡単に取得できる.しかし,実際上は,量子多体系の効果でラインプロファイルが広がる効果を加味する必要があり,例えば,MnのKalpha線はKalpha1,2の強度比が1:2で終わりのはずが,7つや8つのVoigt関数をいれてフィットしないと,正しく装置の分解能を評価できない.いくつか代表的なラインプロファイルのパラメータと簡単な生成方法をpythonでまとめておいた.
その前に、ラインの呼び方には2通りあることを知っておいた方がよい。 Siegbahn と IUPAC という読み方がある、Siegbahn notation など。簡単に考えると、IUPAC は、KL1,KL2,KL3のように数字が大きくなるほど、エネルギーが高くなる。例えば、L1, L2, and L3 が、2S1/2, 2P1/2, and 2P3/2 に対応しているためである。2S1/2の読み方は、[蛍光X線スペクトルの読み方について] (http://www.process.mtl.kyoto-u.ac.jp/pdf/XRF2008.pdf) など参照ください。主量子数nを冒頭の数字、軌道角運動量をアルファベット、全角運動量(spin+orbit)を3つめの数字で表記したものである。
実測スペクトルをモデルフィットする場合は,pythonで複数のvoigt関数にレスポンスを入れてフィットする方法 を参照してほしい.
コード
Lineprofile クラスが,ラインのエネルギー,強度,自然幅,装置の分解能を加味して,Voigt関数を生成する.mymodel クラスの中では,それをrawfuncという関数にして,必要なら低エネルギー側のテイルの計算も行えるようにしている.
#!/usr/bin/env python
__version__= '1.0'
import matplotlib.pyplot as plt
plt.rcParams['font.family'] = 'serif'
import numpy as np
import scipy.special
def mymodel(x,params, consts=[], tailonly = False):
norm,gw,gain,P_tailfrac,P_tailtau,bkg1,bkg2 = params
# norm : nomarlizaion
# gw : sigma of gaussian
# gain : gain of the spectrum
# P_tailfrac : fraction of tail
# P_tailtau : width of the low energy tail
# bkg1 : constant of background
# bkg2 : linearity of background
initparams = [norm,gw,gain,bkg1,bkg2]
def rawfunc(x): # local function, updated when mymodel is called
return Lineprofile(x,initparams,consts=consts)
model_y = smear(rawfunc, x, P_tailfrac, P_tailtau, tailonly=tailonly)
return model_y
def Lineprofile(xval,params,consts=[]):
norm,gw,gain,bkg1,bkg2 = params
# norm : normalization
# gw : sigma of the gaussian
# gain : if gain changes
# consttant facter if needed
prob = (amp * lgamma) / np.sum(amp * lgamma) # probabilites for each lines.
model_y = 0
if len(consts) == 0:
consts = np.ones(len(energy))
else:
consts = consts
for i, (ene,lg,pr,con) in enumerate(zip(energy,lgamma,prob,consts)):
voi = voigt(xval,[ene*gain,lg*0.5,gw])
model_y += norm * con * pr * voi
background = bkg1 * np.ones(len(xval)) + (xval - np.mean(xval)) * bkg2
model_y = model_y + background
# print "bkg1,bkg2 = ", bkg1,bkg2, background
return model_y
def voigt(xval,params):
center,lw,gw = params
# center : center of Lorentzian line
# lw : HWFM of Lorentzian (half-width at half-maximum (HWHM))
# gw : sigma of the gaussian
z = (xval - center + 1j*lw)/(gw * np.sqrt(2.0))
w = scipy.special.wofz(z)
model_y = (w.real)/(gw * np.sqrt(2.0*np.pi))
return model_y
def smear(rawfunc, x, P_tailfrac, P_tailtau, tailonly = False):
if P_tailfrac <= 1e-5:
return rawfunc(x)
dx = x[1] - x[0]
freq = np.fft.rfftfreq(len(x), d=dx)
rawspectrum = rawfunc(x)
ft = np.fft.rfft(rawspectrum)
if tailonly:
ft *= P_tailfrac * (1.0 / (1 - 2j * np.pi * freq * P_tailtau) - 0)
else:
ft += ft * P_tailfrac * (1.0 / (1 - 2j * np.pi * freq * P_tailtau) - 1)
smoothspectrum = np.fft.irfft(ft, n=len(x))
if tailonly:
pass
else:
smoothspectrum[smoothspectrum < 0] = 0
return smoothspectrum
class aline:
def __init__(self,x,y,name):
self.x = x
self.y = y
self.name = name
# global variables
gfwhm = 2
gw = gfwhm / 2.35
norm = 500000.0
gain = 1.0
bkg1 = 1.0
bkg2 = 0.0
P_tailfrac = 1e-6 # no tail
P_tailtau = 10
nbin=1000
ewidth=500
init_params=[norm,gw,gain,P_tailfrac,P_tailtau,bkg1,bkg2]
linelist = []
#################################################################
name="Ti Kalpha"
energy = np.array((4510.918, 4509.954, 4507.763, 4514.002, 4504.910, 4503.088))
lgamma = np.array((1.37, 2.22, 3.75, 1.70, 1.88, 4.49))
amp = np.array((4549, 626, 236, 143, 2034, 54))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Ti KBeta"
energy = np.array((25.37, 30.096, 31.967, 35.59)) + 4900
lgamma = np.array((16.3, 4.25, 0.42, 0.47))
amp = np.array((199, 455, 326, 19.2))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="V Kalpha"
energy = np.array((4952.237, 4950.656, 4948.266, 4955.269, 4944.672, 4943.014))
lgamma = np.array((1.45, 2.00, 1.81, 1.76, 2.94, 3.09))
amp = np.array((25832, 5410, 1536, 956, 12971, 603))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="V KBeta"
energy = np.array((18.19, 24.50, 26.992)) + 5400
lgamma = np.array((18.86, 5.48, 2.499))
amp = np.array((258, 236, 507))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Cr Kalpha"
energy = 5400 + np.array([14.874, 14.099, 12.745, 10.583, 18.304, 5.551, 3.986])
lgamma = np.array([1.457, 1.760, 3.138, 5.149, 1.988, 2.224, 4.4740])
amp = np.array([882, 237, 85, 45, 15, 386, 36])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Cr KBeta"
energy = 5900 + np.array((47.00, 35.31, 46.24, 42.04, 44.93))
lgamma = np.array([1.70, 15.98, 1.90, 6.69, 3.37])
amp = np.array([670, 55, 337, 82, 151])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Mn Kalpha"
energy = 5800 + np.array((98.853, 97.867, 94.829, 96.532, 99.417, 102.712, 87.743, 86.495))
lgamma = np.array([1.715, 2.043, 4.499, 2.663, 0.969, 1.553, 2.361, 4.216])
amp = np.array([790, 264, 68, 96, 71, 10, 372, 100])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Mn KBeta"
energy = 6400 + np.array((90.89, 86.31, 77.73, 90.06, 88.83))
lgamma = np.array((1.83, 9.40, 13.22, 1.81, 2.81))
amp = np.array([608, 109, 77, 397, 176])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Fe Kalpha"
energy = np.array((6404.148, 6403.295, 6400.653, 6402.077, 6391.190, 6389.106, 6390.275))
lgamma = np.array((1.613, 1.965, 4.833, 2.803, 2.487, 2.339, 4.433))
amp = np.array([697, 376, 88, 136, 339, 60, 102])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Fe Kbeta"
energy = np.array((7046.90, 7057.21, 7058.36, 7054.75))
lgamma = np.array((14.17, 3.12, 1.97, 6.38))
amp = np.array([107, 448, 615, 141])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Co Kalpha"
energy = np.array((6930.425, 6929.388, 6927.676, 6930.941, 6915.713, 6914.659, 6913.078))
lgamma = np.array((1.795, 2.695, 4.555, 0.808, 2.406, 2.773, 4.463))
amp = np.array((809, 205, 107, 41, 314, 131, 43))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Co Kbeta"
energy = np.array((7649.60, 7647.83, 7639.87, 7645.49, 7636.21, 7654.13))
lgamma = np.array((3.05, 3.58, 9.78, 4.89, 13.59, 3.79))
amp = np.array((798, 286, 85, 114, 33, 35))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Ni Kalpha"
energy = np.array((7478.281, 7476.529, 7461.131, 7459.874, 7458.029))
lgamma = np.array((2.013, 4.711, 2.674, 3.039, 4.476))
amp = np.array((909, 136, 351, 79, 24))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Ni Kbeta"
energy = np.array((8265.01, 8263.01, 8256.67, 8268.70))
lgamma = np.array((3.76, 4.34, 13.70, 5.18))
amp = np.array((722, 358, 89, 104))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Cu Kalpha"
energy = np.array([8047.8372, 8045.3672, 8027.9935, 8026.5041])
lgamma = np.array([2.285, 3.358, 2.667, 3.571])
amp = np.array([957, 90, 334, 111])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="Cu KBeta"
energy = np.array([8905.532, 8903.109, 8908.462, 8897.387, 8911.39])
lgamma = np.array([3.52, 3.52, 3.55, 8.08, 5.31])
amp = np.array([757, 388, 171, 68, 55])
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="As Kalpha"
energy = np.array((10543.2674,10507.50))
lgamma = np.array((3.08, 3.17))
amp = np.array((1.00, 0.51))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
#################################################################
name="As KBeta"
energy = np.array((11725.73,11719.86))
lgamma = np.array((2.09+2.25, 2.09+2.25))
amp = np.array((0.13, 0.06))
xmin=np.mean(energy) - ewidth
xmax=np.mean(energy) + ewidth
x = np.linspace(xmin,xmax,nbin)
model_y = mymodel(x,init_params)
linelist.append(aline(x,model_y,name))
plt.figure(figsize=(10,8))
plt.title("Line Profiles")
for oneline in linelist:
plt.xlabel("Energy (eV)")
plt.plot(oneline.x, oneline.y, '-', label = oneline.name)
plt.legend(numpoints=1, frameon=False, loc="best")
plt.grid(linestyle='dotted',alpha=0.5)
plt.savefig("linelist.png")
plt.show()
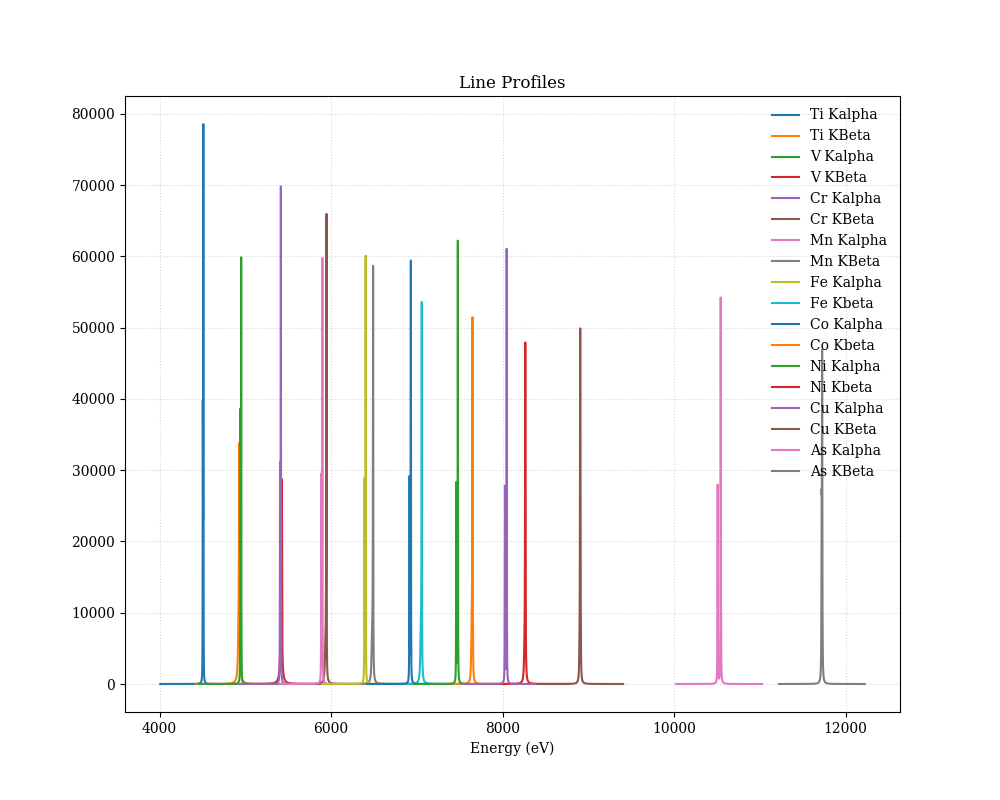
使い方
単純に実行すると,このような図が生成される.
もし,装置の低エネルギー側のテイルを入れたい場合は,P_tailfrac = 1e-6 # no tail の部分を大きな数字にすればよい.
energy,lgamma,amp,という global 変数を用意して,それを書き換えるという邪道な書き方をしているが,正しくは,データベースとなる情報はそもそもコードと一体管理しないほうが良いし,ラインごとにちゃんとしたオブジェクトを生成した方がよい.