本家webのドキュメントやサンプルは不親切なので,わかりやすいデータでやってみた.
まずは準備
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from pystruct.inference import inference_dispatch
内容は,PyStructでHMMを実装と同様に時系列データのノイズ除去.学習には,固定時系列にノイズを乗せた時系列を使う.(固定だから推論しなくてもいいじゃん,というのはさておき)
学習データの作成
n_samples = 500
d = np.array([12, 12, 11, 11, 10, 9, 8, 8, 7, 6, 6, 6, 7, 8, 8, 8, 6,
5, 4, 3, 3, 3, 2, 1, 0, 1, 3, 4, 5, 6, 8, 8, 9, 9,
10, 11, 12, 13, 14, 14, 14, 15, 15, 15, 15])
n_nodes = d.shape[0]
n_states = np.unique(d).shape[0]
n_features = n_states + 1 # add bias
y = np.repeat(d[np.newaxis,:], n_samples, axis=0)
data = y + (np.random.rand(n_samples, n_nodes)-0.5)*5
# negative sign for maximization !
X = np.array( [ [ [ -abs(i-j)**0.1 for j in range(n_states)] for i in dd ] for dd in data] )
# add constant features for bias
X = np.array( [np.hstack((X[i], 0.1*np.ones((X[i].shape[0],1)))) for i in range(X.shape[0])] )
データXは,個数500,時系列の長さ45,状態数・クラス数が16,特徴量数は17(SVMのバイアス分).
サイズの確認
X.shape, y.shape
===
((500, 45, 17), (500, 45))
お決まりのように学習とテストを分割
from sklearn.cross_validation import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=0)
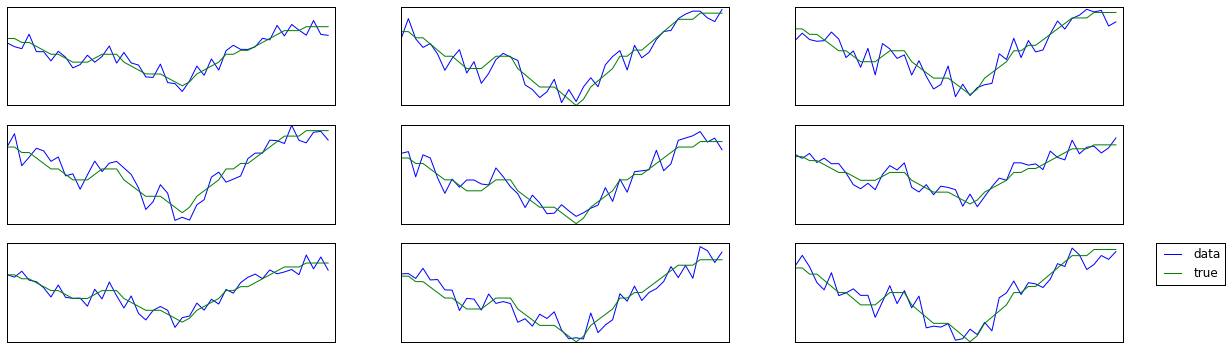
学習データを確認
fig, axes = plt.subplots(3,3, figsize=(20,6))
c=0
for ax in axes.ravel():
ax.plot(data[c], label='data')
ax.plot(y_train[c], label='true')
ax.set_xticks(())
ax.set_yticks(())
c += 1
plt.legend(bbox_to_anchor=(1.1, 1.0), loc=2, borderaxespad=0.)
学習データX(各時刻における特徴)とy(真の固定時系列)を確認のため比較.
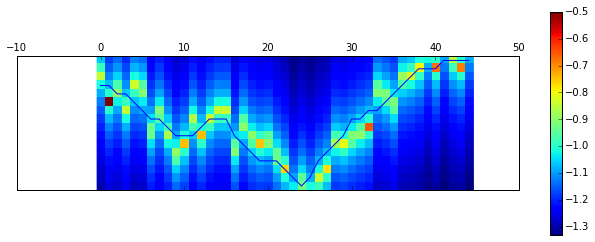
確認
plt.matshow(np.flipud(X_train[0,:,:-1].T)) # remove bias
plt.colorbar()
plt.yticks(())
#plt.show()
plt.plot(15-y_train[0]) # flipud
plt.show()
では学習器の準備.
PyStructのChainCRFの説明にしたがって,FrancWolfeSSVMで学習.
学習器の準備
from pystruct.models import ChainCRF
from pystruct.learners import FrankWolfeSSVM
model = ChainCRF()
ssvm = FrankWolfeSSVM(model=model, C=.1, max_iter=10)
学習!
%%time
ssvm.fit(X_train, y_train)
====
CPU times: user 1.25 s, sys: 17.4 ms, total: 1.27 s
Wall time: 1.3 s
FrankWolfeSSVM(C=0.1, batch_mode=False, check_dual_every=10,
do_averaging=True, line_search=True, logger=None, max_iter=10,
model=ChainCRF(n_states: 16, inference_method: max-product),
n_jobs=1, random_state=None, sample_method='perm',
show_loss_every=0, tol=0.001, verbose=0)
それでは予測スコアは?
ssvm.score(X_test, y_test)
==========
0.56377777777777771
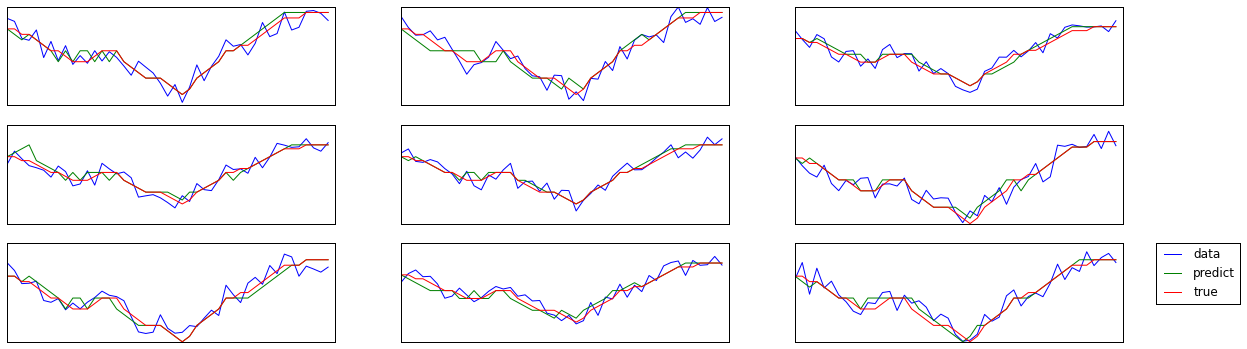
テストに対する予測を確認
X_test_predict = np.array(ssvm.predict(X_test))
fig, axes = plt.subplots(3,3, figsize=(20,6))
shf = np.arange(X_test.shape[0])
np.random.shuffle(shf)
c=0
for ax in axes.ravel():
ax.plot(data[shf[c]], label='data')
ax.plot(X_test_predict[shf[c]], label='predict')
ax.plot(y_test[shf[c]], label='true')
ax.set_xticks(())
ax.set_yticks(())
c += 1
plt.legend(bbox_to_anchor=(1.1, 1.0), loc=2, borderaxespad=0.)
学習されたwを確認
ssvm.w.shape # = n_features * n_states + n_states**2
========
(528,)
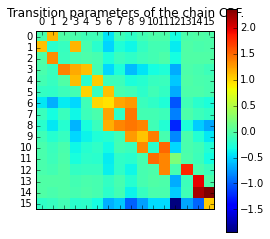
ペアワイズの重みw
plt.matshow(ssvm.w[n_features * n_states:].reshape(n_states, n_states))
plt.title("Transition parameters of the chain CRF.")
plt.xticks(np.arange(n_states))
plt.yticks(np.arange(n_states))
plt.colorbar()
plt.show()
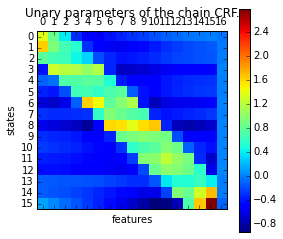
unaryの重みw
plt.matshow(ssvm.w[:n_features * n_states].reshape(n_states,n_features))
plt.title("Unary parameters of the chain CRF.")
plt.yticks(np.arange(n_states))
plt.xticks(np.arange(n_features))
plt.ylabel('states')
plt.xlabel('features')
plt.colorbar()
plt.show()