tldr
KggleのIndian Liver Patient RecordsをLiver Disease Prediction - Data Every Day #04に沿ってやっていきます。
実行環境はGoogle Colaboratorです。
インポート
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn.preprocessing as sp
from sklearn.model_selection import train_test_split
import sklearn.linear_model as slm
import tensorflow as tf
データのダウンロード
Google Driveをマウントします。
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
KaggleのAPIクライアントを初期化し、認証します。
認証情報はGoogle Drive内(/content/drive/My Drive/Colab Notebooks/Kaggle)にkaggle.jsonとして置いてあります。
import os
kaggle_path = "/content/drive/My Drive/Colab Notebooks/Kaggle"
os.environ['KAGGLE_CONFIG_DIR'] = kaggle_path
from kaggle.api.kaggle_api_extended import KaggleApi
api = KaggleApi()
api.authenticate()
Kaggle APIを使ってデータをダウンロードします。
dataset_id = 'uciml/indian-liver-patient-records'
dataset = api.dataset_list_files(dataset_id)
file_name = dataset.files[0].name
file_path = os.path.join(api.get_default_download_dir(), file_name)
file_path
'/content/indian_liver_patient.csv'
api.dataset_download_file(dataset_id, file_name, force=True, quiet=False)
100%|██████████| 23.4k/23.4k [00:00<00:00, 4.49MB/s]
Downloading indian_liver_patient.csv to /content
True
データの読み込み
data = pd.read_csv(file_path)
data
| Age | Gender | Total_Bilirubin | Direct_Bilirubin | Alkaline_Phosphotase | Alamine_Aminotransferase | Aspartate_Aminotransferase | Total_Protiens | Albumin | Albumin_and_Globulin_Ratio | Dataset | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 65 | Female | 0.7 | 0.1 | 187 | 16 | 18 | 6.8 | 3.3 | 0.90 | 1 |
| 1 | 62 | Male | 10.9 | 5.5 | 699 | 64 | 100 | 7.5 | 3.2 | 0.74 | 1 |
| 2 | 62 | Male | 7.3 | 4.1 | 490 | 60 | 68 | 7.0 | 3.3 | 0.89 | 1 |
| 3 | 58 | Male | 1.0 | 0.4 | 182 | 14 | 20 | 6.8 | 3.4 | 1.00 | 1 |
| 4 | 72 | Male | 3.9 | 2.0 | 195 | 27 | 59 | 7.3 | 2.4 | 0.40 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 578 | 60 | Male | 0.5 | 0.1 | 500 | 20 | 34 | 5.9 | 1.6 | 0.37 | 2 |
| 579 | 40 | Male | 0.6 | 0.1 | 98 | 35 | 31 | 6.0 | 3.2 | 1.10 | 1 |
| 580 | 52 | Male | 0.8 | 0.2 | 245 | 48 | 49 | 6.4 | 3.2 | 1.00 | 1 |
| 581 | 31 | Male | 1.3 | 0.5 | 184 | 29 | 32 | 6.8 | 3.4 | 1.00 | 1 |
| 582 | 38 | Male | 1.0 | 0.3 | 216 | 21 | 24 | 7.3 | 4.4 | 1.50 | 2 |
583 rows × 11 columns
下準備
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 583 entries, 0 to 582
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Age 583 non-null int64
1 Gender 583 non-null object
2 Total_Bilirubin 583 non-null float64
3 Direct_Bilirubin 583 non-null float64
4 Alkaline_Phosphotase 583 non-null int64
5 Alamine_Aminotransferase 583 non-null int64
6 Aspartate_Aminotransferase 583 non-null int64
7 Total_Protiens 583 non-null float64
8 Albumin 583 non-null float64
9 Albumin_and_Globulin_Ratio 579 non-null float64
10 Dataset 583 non-null int64
dtypes: float64(5), int64(5), object(1)
memory usage: 50.2+ KB
data.isna().sum()
Age 0
Gender 0
Total_Bilirubin 0
Direct_Bilirubin 0
Alkaline_Phosphotase 0
Alamine_Aminotransferase 0
Aspartate_Aminotransferase 0
Total_Protiens 0
Albumin 0
Albumin_and_Globulin_Ratio 4
Dataset 0
dtype: int64
data['Albumin_and_Globulin_Ratio'] = data['Albumin_and_Globulin_Ratio'].fillna(data['Albumin_and_Globulin_Ratio'].mean())
エンコード
def binary_encode(df, column, positive_value):
df = df.copy()
df[column] = df[column].apply(lambda x: 1 if x == positive_value else 0)
return df
data = binary_encode(data, 'Gender', 'Male')
data = binary_encode(data, 'Dataset', 1)
分割とスケーリング
y = data['Dataset']
X = data.drop('Dataset', axis=1)
scaler = sp.StandardScaler()
X = scaler.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8)
トレーニング
X.shape
(583, 10)
y.sum() / len(y)
0.7135506003430532
model = tf.keras.Sequential([
tf.keras.layers.Dense(64, activation='relu', input_shape=(10,)),
tf.keras.layers.Dense(64, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.summary()
model.compile(
optimizer='adam',
loss='binary_crossentropy',
metrics=[
'accuracy',
tf.keras.metrics.AUC(name='auc'),
],
)
batch_size = 64
epochs = 100
history = model.fit(
X_train,
y_train,
validation_split=0.2,
batch_size=batch_size,
epochs=epochs,
verbose=0,
)
Model: "sequential_3"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_9 (Dense) (None, 64) 704
_________________________________________________________________
dense_10 (Dense) (None, 64) 4160
_________________________________________________________________
dense_11 (Dense) (None, 1) 65
=================================================================
Total params: 4,929
Trainable params: 4,929
Non-trainable params: 0
_________________________________________________________________
結果
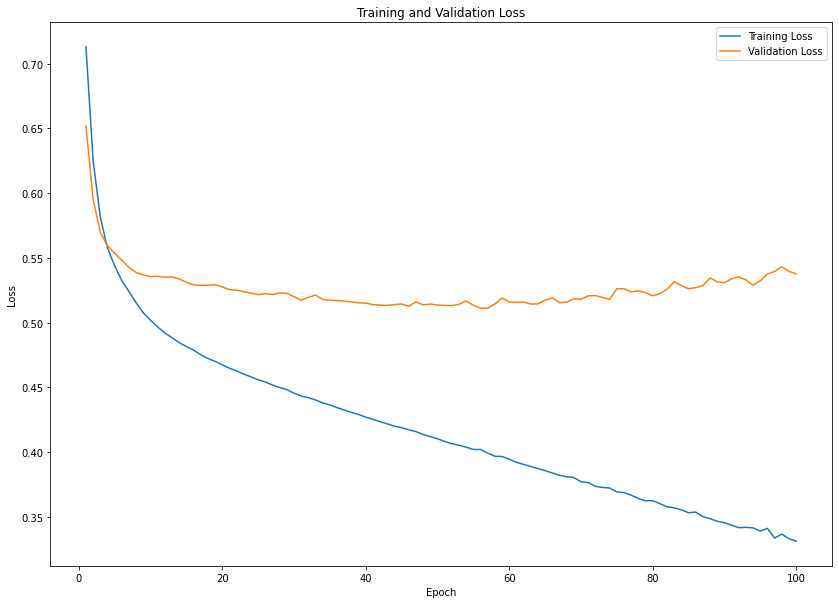
plt.figure(figsize=(14, 10))
epochs_range = range(1, epochs+1)
train_loss = history.history['loss']
val_loss = history.history['val_loss']
plt.plot(epochs_range, train_loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Validation Loss')
plt.title('Training and Validation Loss')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend()
plt.show()
np.argmin(val_loss)
56
model.evaluate(X_test, y_test)
4/4 [==============================] - 0s 4ms/step - loss: 0.6818 - accuracy: 0.6325 - auc: 0.7016
[0.681831419467926, 0.632478654384613, 0.7015877962112427]