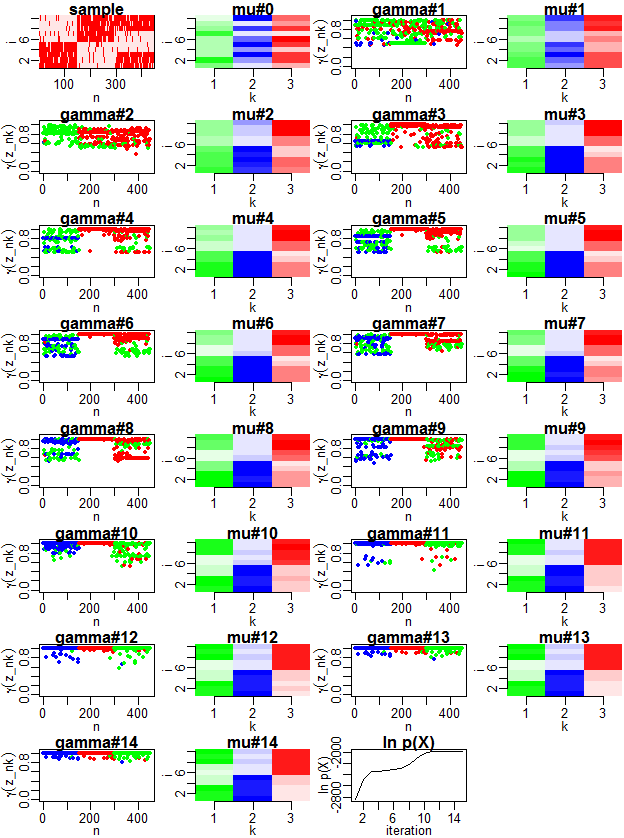
PRML 9.3.3に記載の通り、EMアルゴリズムによって混合ベルヌーイモデルの最尤推定が行われる過程と、対数尤度関数の収束の様子を示します。
10次元2値サンプルを、3種のベルヌーイ分布に従って、150サンプルずつ順に生成し、それらのサンプルに対する対数尤度を最大化します。EMステップ毎に、各サンプルについて、その混合要素のうち、最大の負担率γ(z_nk)の値を、その混合要素を示す色で記します。また、各混合要素について、ベルヌーイ分布のパラメータμkの値を示します。
また、MNISTの手書き数字文字データから生成した784(=28x28)次元2値データについて、数字2,3,4を各150サンプルずつ利用し、それらのサンプルに対する対数尤度を最大化します。
frame()
set.seed(0)
par(mfrow=c(8, 4))
par(mar=c(2.3, 2.5, 1, 0.1))
par(mgp=c(1.3, .5, 0))
MNIST <- F # to load the MNIST handwritten digits
K <- 3
N <- 450
rbern <- function(mu) {
ifelse(runif(length(mu)) < mu, 1, 0)
}
logdbern <- function(x, mu) {
sum(log(mu ^ x * (1 - mu) ^ (1 - x)))
}
logsumexp <- function (x) {
m <- max(x)
m + log(sum(exp(x - m)))
}
if (MNIST) {
# the following code is taken from https://gist.github.com/brendano/39760
load_mnist <- function() {
load_image_file <- function(filename) {
ret = list()
f = file(filename,'rb')
readBin(f,'integer',n=1,size=4,endian='big')
ret$n = readBin(f,'integer',n=1,size=4,endian='big')
nrow = readBin(f,'integer',n=1,size=4,endian='big')
ncol = readBin(f,'integer',n=1,size=4,endian='big')
x = readBin(f,'integer',n=ret$n*nrow*ncol,size=1,signed=F)
ret$x = matrix(x, ncol=nrow*ncol, byrow=T)
close(f)
ret
}
load_label_file <- function(filename) {
f = file(filename,'rb')
readBin(f,'integer',n=1,size=4,endian='big')
n = readBin(f,'integer',n=1,size=4,endian='big')
y = readBin(f,'integer',n=n,size=1,signed=F)
close(f)
y
}
train <<- load_image_file('mnist/train-images-idx3-ubyte')
train$y <<- load_label_file('mnist/train-labels-idx1-ubyte')
}
# the data is available at http://yann.lecun.com/exdb/mnist/
# extract the files in the "mnist" directory
setwd("C:/Users/Public/Documents")
load_mnist()
x <- floor(rbind(
train$x[train$y == 2, ][1:(N / 3), ],
train$x[train$y == 3, ][1:(N / 3), ],
train$x[train$y == 4, ][1:(N / 3), ]) / 128)
D <- ncol(x)
} else {
PHI <- 0.9
PLO <- 0.1
muorg <- matrix(c(
PHI, PHI, PHI, PHI, PHI, PLO, PLO, PLO, PLO, PLO,
PLO, PLO, PLO, PLO, PLO, PHI, PHI, PHI, PHI, PHI,
PHI, PHI, PHI, PLO, PLO, PLO, PLO, PHI, PHI, PHI
), 3, byrow=T)
z <- rep(1:3, each=N/3)
x <- rbern(muorg[z, ])
D <- 10
}
image(1:N, 1:D, x, xlab="n", ylab="i", breaks=seq(0, 1, 0.1),
col=hsv(0, seq(0.1, 1, 0.1), 1))
title("sample")
mu <- matrix(runif(D * K), K, byrow=T)
pz <- rep(1 / K, K)
gamma <- matrix(NA, nrow=N, ncol=K)
likelihood <- numeric()
iteration <- 0
repeat {
cat("mu\n");print(mu)
cat("pi\n");print(pz)
if (!is.na(gamma[1, 1])) {
plot(apply(gamma, 1, max), col=hsv(apply(gamma, 1, which.max) / K, 1, 1),
ylim=c(0, 1.05), pch=20, xlab="n", ylab=expression(gamma(z_nk)))
title(paste0("gamma#", iteration))
}
if (MNIST) {
image(1:28, 1:(28*K), matrix(as.vector(t(mu + rep(0:(K-1), D) + 1.0E-8)), nrow=28)[, (28*K):1],
axes=F, xlab="i", ylab="k",
breaks=seq(0, K, 0.1),
col=outer(seq(0.1, 1, 0.1), (1:K)/K, function(x1, x2) hsv(x2, x1, 1)))
axis(1)
axis(2, at=0:(K-1) * 28 + 14, labels=1:K)
} else {
image(1:K, 1:D, mu + rep(0:(K-1), D) + 1.0E-8,
axes=F, xlab="k", ylab="i",
breaks=seq(0, K, 0.1),
col=outer(seq(0.1, 1, 0.1), (1:K)/K, function(x1, x2) hsv(x2, x1, 1)))
axis(1, at=1:K)
axis(2)
}
title(paste0("mu#", iteration))
# E step
for (n in 1:N) {
pzx <- sapply(
1:K,
function(k) log(pz[k]) + logdbern(x[n, ], mu[k, ])
)
pzx <- pzx - max(pzx)
gamma[n, ] <- exp(pzx) / sum(exp(pzx))
}
# M step
nk <- colSums(gamma)
for (k in 1:K) {
mu[k, ] <- colSums(x * gamma[, k]) / nk[k]
pz[k] <- nk[k] / N
}
# likelihood
likelihood <- c(likelihood, sum(sapply(1:N, function(n)
logsumexp(sapply(
1:K,
function(k)
log(pz[k]) + logdbern(x[n, ], mu[k, ])
))
)))
if (length(likelihood) > 1
&& likelihood[length(likelihood)] - likelihood[length(likelihood) - 1] < 1.0E-2) {
break
}
iteration <- iteration + 1
}
plot(likelihood, type="l", xlab="iteration", ylab="ln p(X)")
title("ln p(X)")