Legacy Autoencoder: https://qiita.com/Rowing0914/items/90e00357248f908d7c6b
Introduction
Since I have finished the reviewing of the foundational paper in autoencoder, I would like to move on to reviewing the research paper about denoising autoencoder.
Looking back the issue arising in autoencoder, it tends to learn identity functions because of something which I will learn in this paper.
Title: Extracting and Composing Robust Features with Denoising
Autoencoders
Author:
Publish Year:
Link: http://www.cs.toronto.edu/~larocheh/publications/icml-2008-denoising-autoencoders.pdf
Denoising Autoencoder
In this model, the difference between legacy autoencoder is the preprocessing step of data.
Initially we use bare inputs denoted $X$, but in this model they have proposed to corrupt the input data certainly and the autoencoder will take as input generated inputs. Other than that, basically same as legacy autoencoder.
Zeroing
They have called the preprocessing procedure as zeroing.
In fact, the step is separated into 3.
- Define V as the length to become 0 in the image(this length is global variable for the entire dataset)
- Calculating the target matrix, which is the $V \times d $ matrix.
- Replace the matrix with 0.
Actually this technique is called "Salt Noise"
from keras.datasets import mnist
import numpy as np
(x_train, _), (x_test, _) = mnist.load_data()
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
x_train = np.reshape(x_train, (len(x_train), 28,28,1))
x_test = np.reshape(x_test, (len(x_test), 28,28,1))
noise_factor = 0.5
x_train_noisy = x_train + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_train.shape)
x_test_noisy = x_test + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_test.shape)
x_train_noisy = np.clip(x_train_noisy, 0., 1.)
x_test_noisy = np.clip(x_test_noisy, 0., 1.)
n = 10
plt.figure(figsize=(20, 4))
for i in range(n):
ax = plt.subplot(1, n, i + 1)
plt.imshow(x_test_noisy[i].reshape(28, 28))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.show()

implementation
from keras.layers import Input, Dense
from keras.models import Model
encoding_dim = 32
input_img = Input(shape=(784, ))
encoded = Dense(encoding_dim, activation='relu')(input_img)
decoded = Dense(784, activation='sigmoid')(encoded)
autoencoder = Model(input_img, decoded)
encoder = Model(input_img, encoded)
encoded_input = Input(shape=(encoding_dim, ))
decoded_layer = autoencoder.layers[-1]
decoder = Model(encoded_input, decoded_layer(encoded_input))
autoencoder.compile(optimizer='adam', loss='binary_crossentropy')
from keras.datasets import mnist
import numpy as np
(x_train, _), (x_test, _) = mnist.load_data()
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
x_train = np.reshape(x_train, (len(x_train), 28,28,1))
x_test = np.reshape(x_test, (len(x_test), 28,28,1))
noise_factor = 0.5
x_train_noisy = x_train + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_train.shape)
x_test_noisy = x_test + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_test.shape)
x_train = np.clip(x_train_noisy, 0., 1.)
x_test = np.clip(x_test_noisy, 0., 1.)
x_train = x_train.reshape((len(x_train), np.prod(x_train.shape[1:])))
x_test = x_test.reshape((len(x_test), np.prod(x_test.shape[1:])))
autoencoder.fit(x_train, x_train, epochs=10, batch_size=256, shuffle=True, validation_data=(x_test, x_test))
# serialize weights to HDF5
autoencoder.save_weights("model.h5")
print("Saved model to disk")
encoded_imgs = encoder.predict(x_test)
decoded_imgs = decoder.predict(encoded_imgs)
import matplotlib.pyplot as plt
n = 10
plt.figure(figsize=(20, 4))
for i in range(n):
ax = plt.subplot(2, n, i+1)
plt.imshow(x_test[i].reshape(28, 28))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
ax = plt.subplot(2, n, i + 1 + n)
plt.imshow(decoded_imgs[i].reshape(28, 28))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.show()
Result Comparison
- Legacy Autoencoder
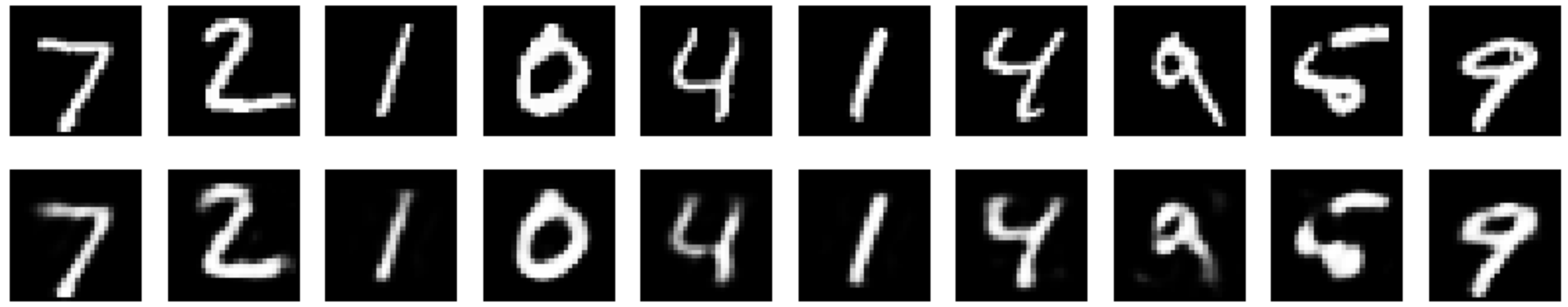 2. Denoising Autoencoder
2. Denoising Autoencoder
