はじめに
pythonの高速化で割とハマったのでここにまとめておく.結論を言うと計算量に応じて最適な高速方法が違うようで,データ数が小さいときはnumbaのjitで最適化したnumpyのcodeがscipyや生のnumpyのcodeより高速であった.つまり,沢山の小さいデータを扱うときはnumpy+jitの方が便利と考えられる.大データを扱う際はscipyやnumpyをjitを無しで使った方が速い.ここでは,相関係数(pearson'r)を計算するprogramで実演しておく.
方法
scipy,numpy,numpy+jit,for-loopを使っただめなcode,for-loopを使っただめなcode+jitのパターンで相関係数を計算するcodeを書いた(codeは末尾に記した).各codeの実行時間を%timeitで計測した.相関を計測する二つのデータはnp.random.randでdatasizeを決めて下記のように取得したもの.
datasize=10
dataA=np.random.rand(datasize)
dataB=np.random.rand(datasize)
実行時間をデータの大きさ(datasize)に対してplotした.
環境はanaconda2.5+python3.6.OSはmacOS High Sierra.
結果
- データのサイズが小さいとき(<10^4)はjitが早い.for-loopのある駄目なcodeでもjitを利用するとscipyより速い!!
- データのサイズが大きいとき(>10^4)は生のscipy or numpyが早い.
- for-loopを使っただめなcodeはjitなしでは(勿論)悲惨.
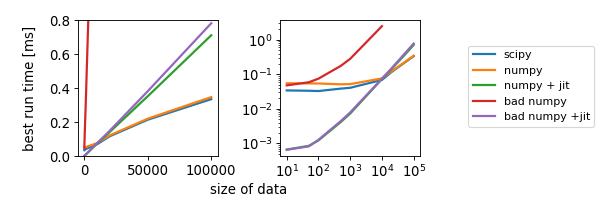
(緑(numpy+jit)と紫(bad numpy+jit)は殆ど重なっている)
(橙(numpy)と青(scipy)も殆ど重なっている)
おわりに
はじめに述べたとおりデータ数が小さいときはjitで最適化したnumpyが生のnumpyのcodeより高速である.
今回は相関係数でしか試していないので結論の一般性については吟味が必要かもしれないが,結論自体は自然な気もする.
##code
import numpy as np
import scipy
from scipy import stats
import numba
# scipy only
def calc_with_scipy(data1,data2):
return scipy.stats.pearsonr(data1,data2)[0]
# numpy only
def calc_with_numpy_plain(data1,data2):
ave1=np.mean(data1)
ave2=np.mean(data2)
std1=np.std(data1)
std2=np.std(data2)
cross_term=data1@data2
cross_term/=len(data1)
cross_term-=ave1*ave2
return cross_term/(std1*std2)
# numpy + jit
@numba.jit
def calc_with_numpy_jit(data1,data2):
ave1=np.mean(data1)
ave2=np.mean(data2)
std1=np.std(data1)
std2=np.std(data2)
cross_term=data1@data2
cross_term/=len(data1)
cross_term-=ave1*ave2
return cross_term/(std1*std2)
# 不必要なloopを使っただめなcode (bad numpy)
def calc_with_numpy_loop_plain(data1,data2):
ave1=np.mean(data1)
ave2=np.mean(data2)
std1=np.std(data1)
std2=np.std(data2)
cross_term=0
for index in range(len(data1)):
cross_term+=data1[index]*data2[index]
cross_term/=len(data1)
cross_term-=ave1*ave2
return cross_term/(std1*std2)
# 不必要なloopを使っただめなcodeをjitで高速化 (bad numpy + jit)
@numba.jit
def calc_with_numpy_loop_jit(data1,data2):
ave1=np.mean(data1)
ave2=np.mean(data2)
std1=np.std(data1)
std2=np.std(data2)
cross_term=0
for index in range(len(data1)):
cross_term+=data1[index]*data2[index]
cross_term/=len(data1)
cross_term-=ave1*ave2
return cross_term/(std1*std2)
#計測用
looplist=(10,50,100,500,1000,10000,20000,50000,100000)
ave_logsA=[]
ave_logsB=[]
ave_logsC=[]
ave_logsD=[]
ave_logsE=[]
best_logsA=[]
best_logsB=[]
best_logsC=[]
best_logsD=[]
best_logsE=[]
#計測開始
for each in looplist:
dataA=np.random.rand(each)
dataB=np.random.rand(each)
log = %timeit -n 10 -r 401 -o calc_with_scipy(dataA,dataB)
best_logsA.append(log.best)
ave_logsA.append(log.average)
log = %timeit -n 10 -r 401 -o calc_with_numpy_plain(dataA,dataB)
best_logsB.append(log.best)
ave_logsB.append(log.average)
log = %timeit -n 10 -r 401 -o calc_with_numpy_jit(dataA,dataB)
best_logsC.append(log.best)
ave_logsC.append(log.average)
# too slow to test large data set.
if each <10001:
log = %timeit -n 10 -r 401 -o calc_with_numpy_loop_plain(dataA,dataB)
best_logsD.append(log.best)
ave_logsD.append(log.average)
log = %timeit -n 10 -r 401 -o calc_with_numpy_loop_jit(dataA,dataB)
best_logsE.append(log.best)
ave_logsE.append(log.average)
#plot
import matplotlib.pyplot as plt
%matplotlib inline
fontsize=12
fig = plt.figure(figsize=(7.5,2.5))
fig.subplots_adjust(left=0.13, bottom=0.22, right=0.70, top=0.90, wspace=0.45, hspace=None)
fig.text((0.13+0.70)/2.0, 0.05, 'size of data', ha='center', va='center', fontsize = fontsize)
fig.text(0.05,(0.22+0.90)/2., 'best run time [ms]', ha='center', va='center', rotation='vertical', fontsize = fontsize)
ax1 = fig.add_subplot(121)
ax1.set_ylim([0,0.8])
ax1.tick_params(labelsize = fontsize)
strA="scipy"
strB="numpy"
strC="numpy + jit"
strD="bad numpy"
strE="bad numpy +jit"
ax1.plot(looplist,np.array(best_logsA)*1e3, lw=2, label=strA)
ax1.plot(looplist,np.array(best_logsB)*1e3, lw=2, label=strB)
ax1.plot(looplist,np.array(best_logsC)*1e3, lw=2, label=strC)
ax1.plot(looplist[:6],np.array(best_logsD[:6])*1e3, lw=2, label=strD)
ax1.plot(looplist,np.array(best_logsE)*1e3, lw=2,label=strE)
ax1.set_xticks([0,50000,100000])
fig.legend(bbox_to_anchor=(0.5, 0.7, 0.5, .100) ,fontsize=(fontsize-2))
ax2 = fig.add_subplot(122)
ax2.tick_params(labelsize = fontsize)
ax2.set_xscale("log")
ax2.set_yscale("log")
ax2.set_xticks([10,100,1000,10000,100000])
ax2.plot(looplist,np.array(best_logsA)*1e3, lw=2, label=strA)
ax2.plot(looplist,np.array(best_logsB)*1e3, lw=2, label=strB)
ax2.plot(looplist,np.array(best_logsC)*1e3, lw=2, label=strC)
ax2.plot(looplist[:6],np.array(best_logsD[:6])*1e3, lw=2, label=strD)
ax2.plot(looplist,np.array(best_logsE)*1e3, lw=2,label=strE)
fig.show()
fig.savefig("./runtime_vs_ndata.png",dpi=80)