Reference
TGS Salt Identification Challenge
Library
import os
from random import randint
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# plt.style.use('seaborn-white')
import seaborn as sns
# sns.set_style("white")
from tqdm import tqdm_notebook
from sklearn.model_selection import train_test_split
from skimage.transform import resize
from keras.preprocessing.image import load_img
from keras import Model
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from keras.models import load_model
from keras.optimizers import Adam
from keras.utils.vis_utils import plot_model
from keras.preprocessing.image import ImageDataGenerator
from keras.layers import Input, Conv2D, Conv2DTranspose, MaxPooling2D
from keras.layers import Activation, BatchNormalization, Dropout
from keras.layers import Add, concatenate, RepeatVector, Reshape
from keras import backend as K
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
Data
img_size_ori = 101
img_size_target = 128
def upsample(img):
if img_size_ori == img_size_target:
return img
return resize(img, (img_size_target, img_size_target), mode='constant', preserve_range=True)
def downsample(img):
if img_size_ori == img_size_target:
return img
return resize(img, (img_size_ori, img_size_ori), mode='constant', preserve_range=True)
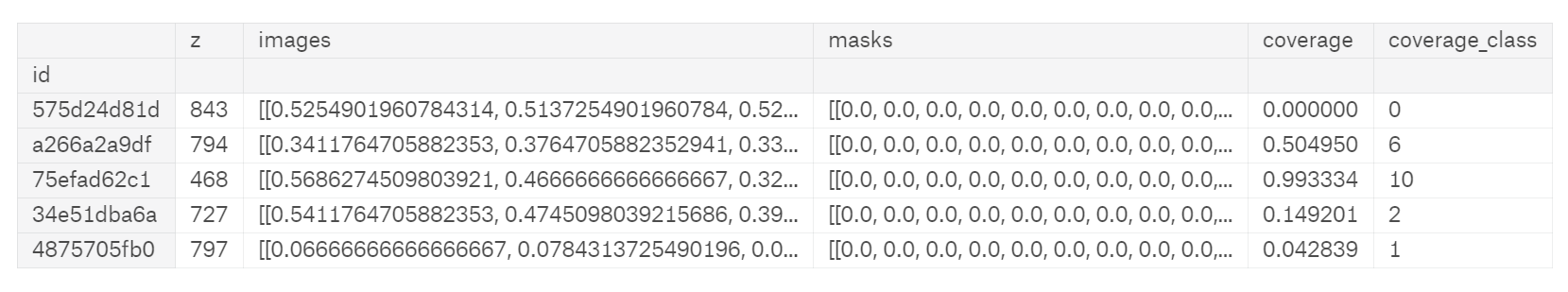
train_df = pd.read_csv("../input/train.csv", index_col="id", usecols=[0])
depths_df = pd.read_csv("../input/depths.csv", index_col="id")
train_df = train_df.join(depths_df)
train_df["images"] = [np.array(load_img("../input/train/images/{}.png".format(idx), grayscale=True)) / 255 for idx in tqdm_notebook(train_df.index)]
train_df["masks"] = [np.array(load_img("../input/train/masks/{}.png".format(idx), grayscale=True)) / 255 for idx in tqdm_notebook(train_df.index)]
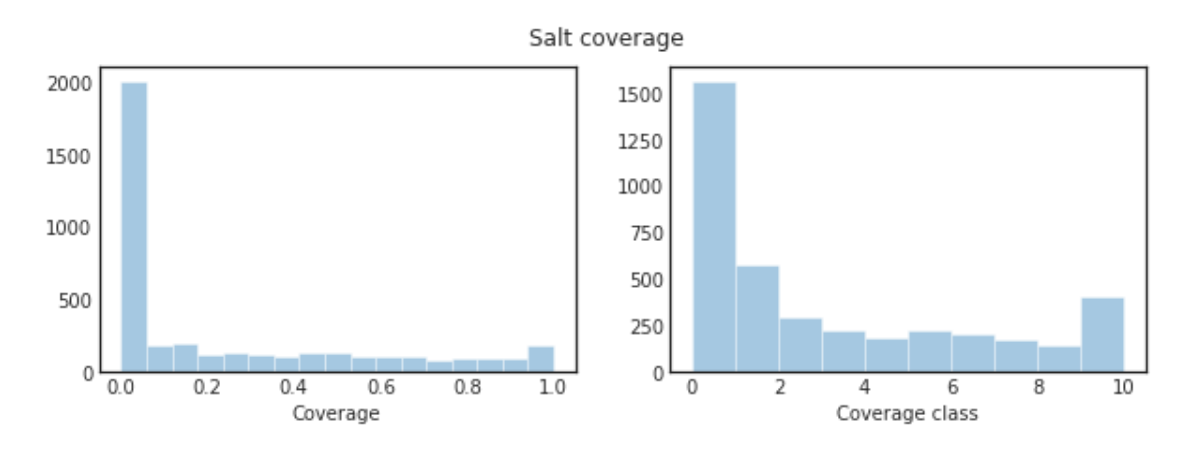
train_df["coverage"] = train_df.masks.map(np.sum) / pow(img_size_ori, 2)
def cov_to_class(val):
for i in range(0, 11):
if val * 10 <= i :
return i
train_df["coverage_class"] = train_df.coverage.map(cov_to_class)
train_df.head()
fig, axs = plt.subplots(1, 2, figsize=(10,3))
sns.distplot(train_df.coverage, kde=False, ax=axs[0])
sns.distplot(train_df.coverage_class, bins=10, kde=False, ax=axs[1])
plt.suptitle("Salt coverage")
axs[0].set_xlabel("Coverage")
axs[1].set_xlabel("Coverage class")
plt.show()
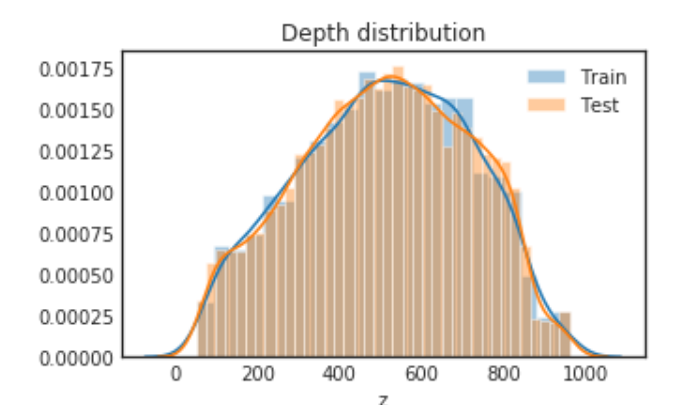
plt.figure(figsize= (5, 3))
sns.distplot(train_df.z, label="Train")
sns.distplot(test_df.z, label="Test")
plt.legend()
plt.title("Depth distribution")
plt.show()
ids_train, ids_valid, x_train, x_valid, y_train, y_valid, cov_train, cov_test, depth_train, depth_test = train_test_split(
train_df.index.values,
np.array(train_df.images.map(upsample).tolist()).reshape(-1, img_size_target, img_size_target, 1),
np.array(train_df.masks.map(upsample).tolist()).reshape(-1, img_size_target, img_size_target, 1),
train_df.coverage.values,
train_df.z.values,
test_size=0.2, stratify=train_df.coverage_class, random_state=1337)
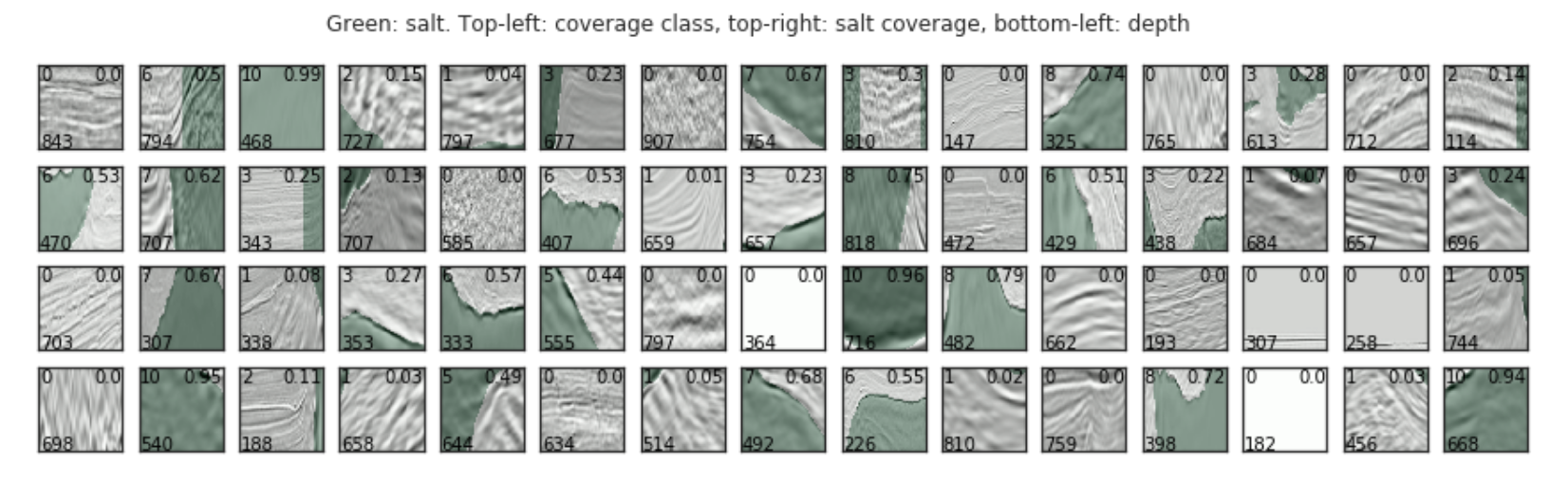
max_images = 60
grid_width = 15
grid_height = int(max_images / grid_width)
plt.figure(figsize=(10, 7))
fig, axs = plt.subplots(grid_height, grid_width, figsize=(grid_width, grid_height))
for i, idx in enumerate(train_df.index[:max_images]):
img = train_df.loc[idx].images
mask = train_df.loc[idx].masks
ax = axs[int(i / grid_width), i % grid_width]
ax.imshow(img, cmap="Greys")
ax.imshow(mask, alpha=0.3, cmap="Greens")
ax.text(1, img_size_ori-1, train_df.loc[idx].z, color="black")
ax.text(img_size_ori - 1, 1, round(train_df.loc[idx].coverage, 2), color="black", ha="right", va="top")
ax.text(1, 1, train_df.loc[idx].coverage_class, color="black", ha="left", va="top")
ax.set_yticklabels([])
ax.set_xticklabels([])
plt.suptitle("Green: salt. Top-left: coverage class, top-right: salt coverage, bottom-left: depth")
plt.show()
Data Augmentation
x_train = np.append(x_train, [np.fliplr(x) for x in x_train], axis=0)
y_train = np.append(y_train, [np.fliplr(x) for x in y_train], axis=0)
Model 1
def build_model(input_layer, start_neurons):
# 128 -> 64
conv1 = Conv2D(start_neurons * 1, (3, 3), activation="relu", padding="same")(input_layer)
conv1 = Conv2D(start_neurons * 1, (3, 3), activation="relu", padding="same")(conv1)
pool1 = MaxPooling2D((2, 2))(conv1)
pool1 = Dropout(0.25)(pool1)
# 64 -> 32
conv2 = Conv2D(start_neurons * 2, (3, 3), activation="relu", padding="same")(pool1)
conv2 = Conv2D(start_neurons * 2, (3, 3), activation="relu", padding="same")(conv2)
pool2 = MaxPooling2D((2, 2))(conv2)
pool2 = Dropout(0.5)(pool2)
# 32 -> 16
conv3 = Conv2D(start_neurons * 4, (3, 3), activation="relu", padding="same")(pool2)
conv3 = Conv2D(start_neurons * 4, (3, 3), activation="relu", padding="same")(conv3)
pool3 = MaxPooling2D((2, 2))(conv3)
pool3 = Dropout(0.5)(pool3)
# 16 -> 8
conv4 = Conv2D(start_neurons * 8, (3, 3), activation="relu", padding="same")(pool3)
conv4 = Conv2D(start_neurons * 8, (3, 3), activation="relu", padding="same")(conv4)
pool4 = MaxPooling2D((2, 2))(conv4)
pool4 = Dropout(0.5)(pool4)
# Middle
convm = Conv2D(start_neurons * 16, (3, 3), activation="relu", padding="same")(pool4)
convm = Conv2D(start_neurons * 16, (3, 3), activation="relu", padding="same")(convm)
# 8 -> 16
deconv4 = Conv2DTranspose(start_neurons * 8, (3, 3), strides=(2, 2), padding="same")(convm)
uconv4 = concatenate([deconv4, conv4])
uconv4 = Dropout(0.5)(uconv4)
uconv4 = Conv2D(start_neurons * 8, (3, 3), activation="relu", padding="same")(uconv4)
uconv4 = Conv2D(start_neurons * 8, (3, 3), activation="relu", padding="same")(uconv4)
# 16 -> 32
deconv3 = Conv2DTranspose(start_neurons * 4, (3, 3), strides=(2, 2), padding="same")(uconv4)
uconv3 = concatenate([deconv3, conv3])
uconv3 = Dropout(0.5)(uconv3)
uconv3 = Conv2D(start_neurons * 4, (3, 3), activation="relu", padding="same")(uconv3)
uconv3 = Conv2D(start_neurons * 4, (3, 3), activation="relu", padding="same")(uconv3)
# 32 -> 64
deconv2 = Conv2DTranspose(start_neurons * 2, (3, 3), strides=(2, 2), padding="same")(uconv3)
uconv2 = concatenate([deconv2, conv2])
uconv2 = Dropout(0.5)(uconv2)
uconv2 = Conv2D(start_neurons * 2, (3, 3), activation="relu", padding="same")(uconv2)
uconv2 = Conv2D(start_neurons * 2, (3, 3), activation="relu", padding="same")(uconv2)
# 64 -> 128
deconv1 = Conv2DTranspose(start_neurons * 1, (3, 3), strides=(2, 2), padding="same")(uconv2)
uconv1 = concatenate([deconv1, conv1])
uconv1 = Dropout(0.5)(uconv1)
uconv1 = Conv2D(start_neurons * 1, (3, 3), activation="relu", padding="same")(uconv1)
uconv1 = Conv2D(start_neurons * 1, (3, 3), activation="relu", padding="same")(uconv1)
#uconv1 = Dropout(0.5)(uconv1)
output_layer = Conv2D(1, (1,1), padding="same", activation="sigmoid")(uconv1)
return output_layer
input_layer = Input((img_size_target, img_size_target, 1))
output_layer = build_model(input_layer, 16)
model = Model(input_layer, output_layer)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
early_stopping = EarlyStopping(patience=10, verbose=1)
# model_checkpoint = ModelCheckpoint("./keras.model", save_best_only=True, verbose=1)
reduce_lr = ReduceLROnPlateau(factor=0.1, patience=5, min_lr=0.00001, verbose=1)
epochs = 20
batch_size = 32
history = model.fit(x_train, y_train,
validation_data=[x_valid, y_valid],
epochs=epochs,
batch_size=batch_size,
#callbacks=[early_stopping, model_checkpoint, reduce_lr],
callbacks=[early_stopping, reduce_lr])
model = load_model("./keras.model")
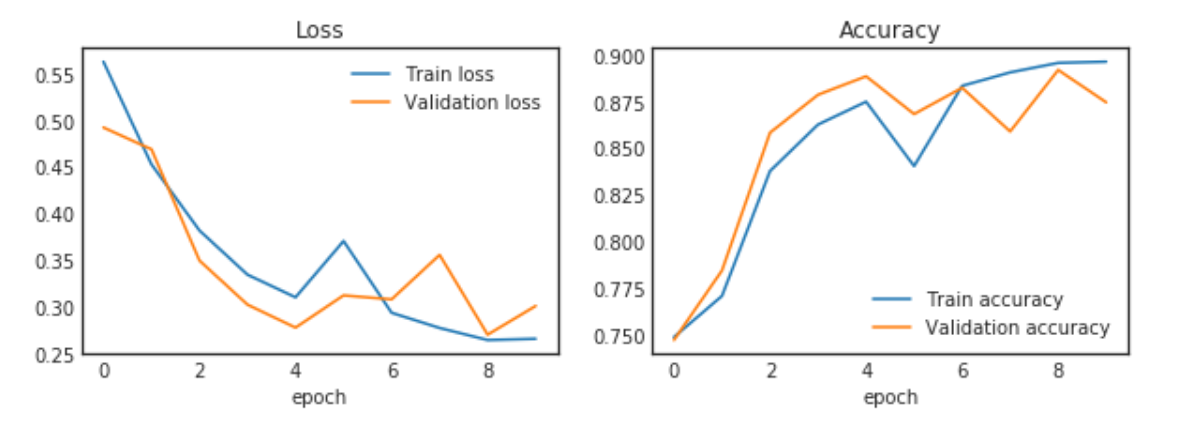
fig, (ax_loss, ax_acc) = plt.subplots(1, 2, figsize=(10,3))
ax_loss.plot(history.epoch, history.history["loss"], label="Train loss")
ax_loss.plot(history.epoch, history.history["val_loss"], label="Validation loss")
ax_loss.set_title('Loss')
ax_loss.set_xlabel('epoch')
ax_loss.legend(loc='best')
ax_acc.plot(history.epoch, history.history["acc"], label="Train accuracy")
ax_acc.plot(history.epoch, history.history["val_acc"], label="Validation accuracy")
ax_acc.set_title('Accuracy')
ax_acc.set_xlabel('epoch')
ax_acc.legend(loc='best')
plt.show()
preds_valid = model.predict(x_valid).reshape(-1, img_size_target, img_size_target)
preds_valid = np.array([downsample(x) for x in preds_valid])
y_valid_ori = np.array([train_df.loc[idx].masks for idx in ids_valid])
max_images = 60
grid_width = 15
grid_height = int(max_images / grid_width)
plt.figure(figsize=(10, 7))
fig, axs = plt.subplots(grid_height, grid_width, figsize=(grid_width, grid_height))
for i, idx in enumerate(ids_valid[:max_images]):
img = train_df.loc[idx].images
mask = train_df.loc[idx].masks
pred = preds_valid[i]
ax = axs[int(i / grid_width), i % grid_width]
ax.imshow(img, cmap="Greys")
ax.imshow(mask, alpha=0.3, cmap="Greens")
ax.imshow(pred, alpha=0.3, cmap="OrRd")
ax.text(1, img_size_ori-1, train_df.loc[idx].z, color="black")
ax.text(img_size_ori - 1, 1, round(train_df.loc[idx].coverage, 2), color="black", ha="right", va="top")
ax.text(1, 1, train_df.loc[idx].coverage_class, color="black", ha="left", va="top")
ax.set_yticklabels([])
ax.set_xticklabels([])
plt.suptitle("Green: salt, Red: prediction. Top-left: coverage class, top-right: salt coverage, bottom-left: depth")
plt.show()
def iou_metric(y_true_in, y_pred_in, print_table=False):
labels = y_true_in
y_pred = y_pred_in
true_objects = 2
pred_objects = 2
intersection = np.histogram2d(labels.flatten(), y_pred.flatten(), bins=(true_objects, pred_objects))[0]
# Compute areas (needed for finding the union between all objects)
area_true = np.histogram(labels, bins = true_objects)[0]
area_pred = np.histogram(y_pred, bins = pred_objects)[0]
area_true = np.expand_dims(area_true, -1)
area_pred = np.expand_dims(area_pred, 0)
# Compute union
union = area_true + area_pred - intersection
# Exclude background from the analysis
intersection = intersection[1:,1:]
union = union[1:,1:]
union[union == 0] = 1e-9
# Compute the intersection over union
iou = intersection / union
# Precision helper function
def precision_at(threshold, iou):
matches = iou > threshold
true_positives = np.sum(matches, axis=1) == 1 # Correct objects
false_positives = np.sum(matches, axis=0) == 0 # Missed objects
false_negatives = np.sum(matches, axis=1) == 0 # Extra objects
tp, fp, fn = np.sum(true_positives), np.sum(false_positives), np.sum(false_negatives)
return tp, fp, fn
# Loop over IoU thresholds
prec = []
if print_table:
print("Thresh\tTP\tFP\tFN\tPrec.")
for t in np.arange(0.5, 1.0, 0.05):
tp, fp, fn = precision_at(t, iou)
if (tp + fp + fn) > 0:
p = tp / (tp + fp + fn)
else:
p = 0
if print_table:
print("{:1.3f}\t{}\t{}\t{}\t{:1.3f}".format(t, tp, fp, fn, p))
prec.append(p)
if print_table:
print("AP\t-\t-\t-\t{:1.3f}".format(np.mean(prec)))
return np.mean(prec)
def iou_metric_batch(y_true_in, y_pred_in):
batch_size = y_true_in.shape[0]
metric = []
for batch in range(batch_size):
value = iou_metric(y_true_in[batch], y_pred_in[batch])
metric.append(value)
return np.mean(metric)
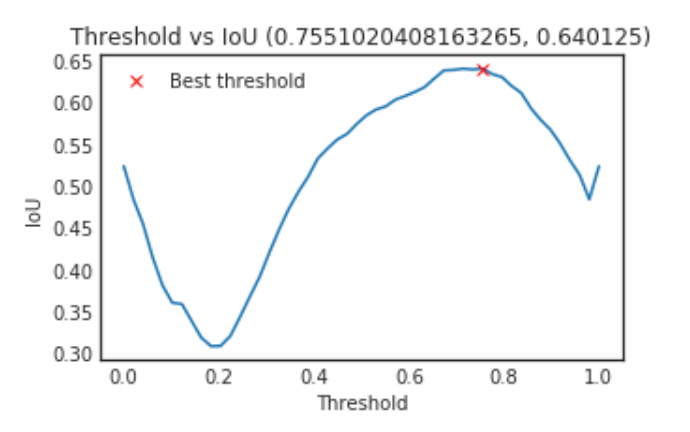
thresholds = np.linspace(0, 1, 50)
ious = np.array([iou_metric_batch(y_valid_ori, np.int32(preds_valid > threshold)) for threshold in tqdm_notebook(thresholds)])
threshold_best_index = np.argmax(ious[9:-10]) + 9
iou_best = ious[threshold_best_index]
threshold_best = thresholds[threshold_best_index]
plt.figure(figsize= (5, 3))
plt.plot(thresholds, ious)
plt.plot(threshold_best, iou_best, "xr", label="Best threshold")
plt.xlabel("Threshold")
plt.ylabel("IoU")
plt.title("Threshold vs IoU ({}, {})".format(threshold_best, iou_best))
plt.legend()
plt.show()
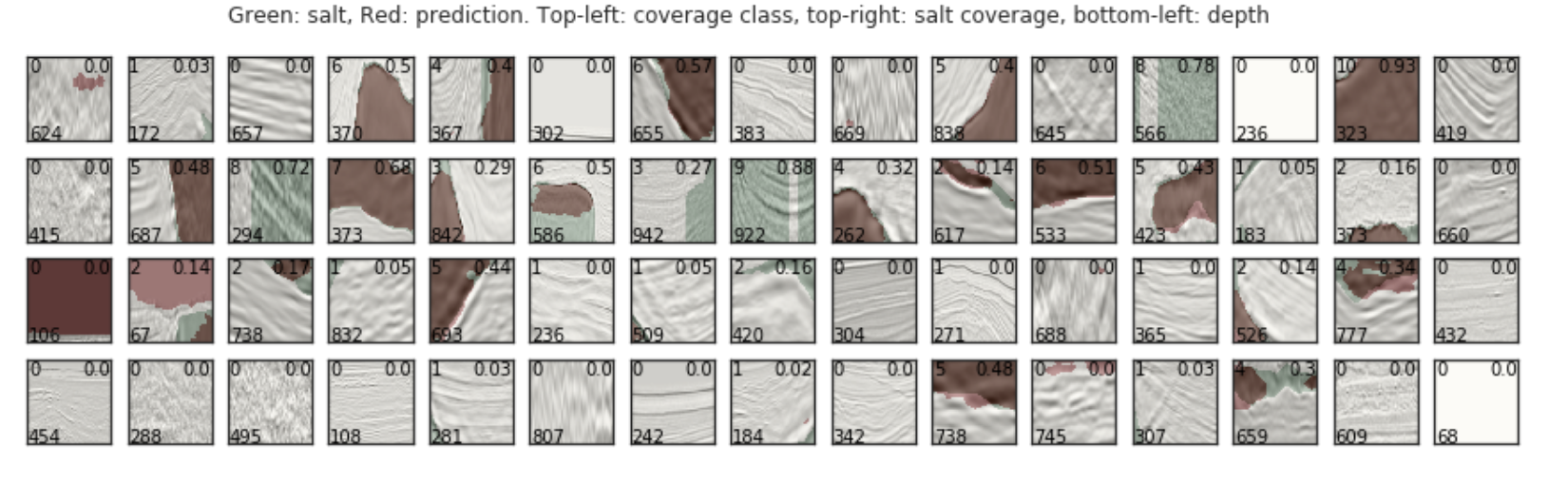
max_images = 60
grid_width = 15
grid_height = int(max_images / grid_width)
fig, axs = plt.subplots(grid_height, grid_width, figsize=(grid_width, grid_height))
plt.figure(figsize=(10, 7))
for i, idx in enumerate(ids_valid[:max_images]):
img = train_df.loc[idx].images
mask = train_df.loc[idx].masks
pred = preds_valid[i]
ax = axs[int(i / grid_width), i % grid_width]
ax.imshow(img, cmap="Greys")
ax.imshow(mask, alpha=0.3, cmap="Greens")
ax.imshow(np.array(np.round(pred > threshold_best), dtype=np.float32), alpha=0.3, cmap="OrRd")
ax.text(1, img_size_ori-1, train_df.loc[idx].z, color="black")
ax.text(img_size_ori - 1, 1, round(train_df.loc[idx].coverage, 2), color="black", ha="right", va="top")
ax.text(1, 1, train_df.loc[idx].coverage_class, color="black", ha="left", va="top")
ax.set_yticklabels([])
ax.set_xticklabels([])
plt.suptitle("Green: salt, Red: prediction. Top-left: coverage class, top-right: salt coverage, bottom-left: depth")
plt.show()
Model 2
def BatchActivate(x):
x = BatchNormalization()(x)
x = Activation('relu')(x)
return x
def convolution_block(x, filters, size, strides=(1,1), padding='same', activation=True):
x = Conv2D(filters, size, strides=strides, padding=padding)(x)
if activation == True:
x = BatchActivate(x)
return x
def residual_block(blockInput, num_filters=16, batch_activate = False):
x = BatchActivate(blockInput)
x = convolution_block(x, num_filters, (3,3) )
x = convolution_block(x, num_filters, (3,3), activation=False)
x = Add()([x, blockInput])
if batch_activate:
x = BatchActivate(x)
return x
def build_model(input_layer, start_neurons, DropoutRatio = 0.5):
# 101 -> 50 (128 -> 64)
conv1 = Conv2D(start_neurons * 1, (3, 3), activation=None, padding="same")(input_layer)
conv1 = residual_block(conv1,start_neurons * 1)
conv1 = residual_block(conv1,start_neurons * 1, True)
pool1 = MaxPooling2D((2, 2))(conv1)
pool1 = Dropout(DropoutRatio/2)(pool1)
# 50 -> 25 (64 -> 32)
conv2 = Conv2D(start_neurons * 2, (3, 3), activation=None, padding="same")(pool1)
conv2 = residual_block(conv2,start_neurons * 2)
conv2 = residual_block(conv2,start_neurons * 2, True)
pool2 = MaxPooling2D((2, 2))(conv2)
pool2 = Dropout(DropoutRatio)(pool2)
# 25 -> 12 (32 -> 16)
conv3 = Conv2D(start_neurons * 4, (3, 3), activation=None, padding="same")(pool2)
conv3 = residual_block(conv3,start_neurons * 4)
conv3 = residual_block(conv3,start_neurons * 4, True)
pool3 = MaxPooling2D((2, 2))(conv3)
pool3 = Dropout(DropoutRatio)(pool3)
# 12 -> 6 (16 -> 8)
conv4 = Conv2D(start_neurons * 8, (3, 3), activation=None, padding="same")(pool3)
conv4 = residual_block(conv4,start_neurons * 8)
conv4 = residual_block(conv4,start_neurons * 8, True)
pool4 = MaxPooling2D((2, 2))(conv4)
pool4 = Dropout(DropoutRatio)(pool4)
# Middle
convm = Conv2D(start_neurons * 16, (3, 3), activation=None, padding="same")(pool4)
convm = residual_block(convm,start_neurons * 16)
convm = residual_block(convm,start_neurons * 16, True)
# 6 -> 12 (8 -> 16)
deconv4 = Conv2DTranspose(start_neurons * 8, (3, 3), strides=(2, 2), padding="same")(convm)
uconv4 = concatenate([deconv4, conv4])
uconv4 = Dropout(DropoutRatio)(uconv4)
uconv4 = Conv2D(start_neurons * 8, (3, 3), activation=None, padding="same")(uconv4)
uconv4 = residual_block(uconv4,start_neurons * 8)
uconv4 = residual_block(uconv4,start_neurons * 8, True)
# 12 -> 25 (16 -> 32)
#deconv3 = Conv2DTranspose(start_neurons * 4, (3, 3), strides=(2, 2), padding="valid")(uconv4)
deconv3 = Conv2DTranspose(start_neurons * 4, (3, 3), strides=(2, 2), padding="same")(uconv4)
uconv3 = concatenate([deconv3, conv3])
uconv3 = Dropout(DropoutRatio)(uconv3)
uconv3 = Conv2D(start_neurons * 4, (3, 3), activation=None, padding="same")(uconv3)
uconv3 = residual_block(uconv3,start_neurons * 4)
uconv3 = residual_block(uconv3,start_neurons * 4, True)
# 25 -> 50 (32 -> 64)
deconv2 = Conv2DTranspose(start_neurons * 2, (3, 3), strides=(2, 2), padding="same")(uconv3)
uconv2 = concatenate([deconv2, conv2])
uconv2 = Dropout(DropoutRatio)(uconv2)
uconv2 = Conv2D(start_neurons * 2, (3, 3), activation=None, padding="same")(uconv2)
uconv2 = residual_block(uconv2,start_neurons * 2)
uconv2 = residual_block(uconv2,start_neurons * 2, True)
# 50 -> 101 (64 -> 128)
#deconv1 = Conv2DTranspose(start_neurons * 1, (3, 3), strides=(2, 2), padding="valid")(uconv2)
deconv1 = Conv2DTranspose(start_neurons * 1, (3, 3), strides=(2, 2), padding="same")(uconv2)
uconv1 = concatenate([deconv1, conv1])
uconv1 = Dropout(DropoutRatio)(uconv1)
uconv1 = Conv2D(start_neurons * 1, (3, 3), activation=None, padding="same")(uconv1)
uconv1 = residual_block(uconv1,start_neurons * 1)
uconv1 = residual_block(uconv1,start_neurons * 1, True)
#uconv1 = Dropout(DropoutRatio/2)(uconv1)
#output_layer = Conv2D(1, (1,1), padding="same", activation="sigmoid")(uconv1)
output_layer_noActi = Conv2D(1, (1,1), padding="same", activation=None)(uconv1)
output_layer = Activation('sigmoid')(output_layer_noActi)
return output_layer
input_layer = Input((img_size_target, img_size_target, 1))
output_layer = build_model(input_layer, 16 , 0.5)
model2 = Model(input_layer, output_layer)
model2.compile(loss="binary_crossentropy", optimizer='adam', metrics=['accuracy'])
model2.summary()
early_stopping = EarlyStopping(patience=5, verbose=1)
# model_checkpoint = ModelCheckpoint("./keras.model", save_best_only=True, verbose=1)
reduce_lr = ReduceLROnPlateau(factor=0.1, patience=5, min_lr=0.00001, verbose=1)
epochs = 10
batch_size = 32
history = model2.fit(x_train, y_train,
validation_data=[x_valid, y_valid],
epochs=epochs,
batch_size=batch_size,
#callbacks=[early_stopping, model_checkpoint, reduce_lr],
callbacks=[early_stopping, reduce_lr])
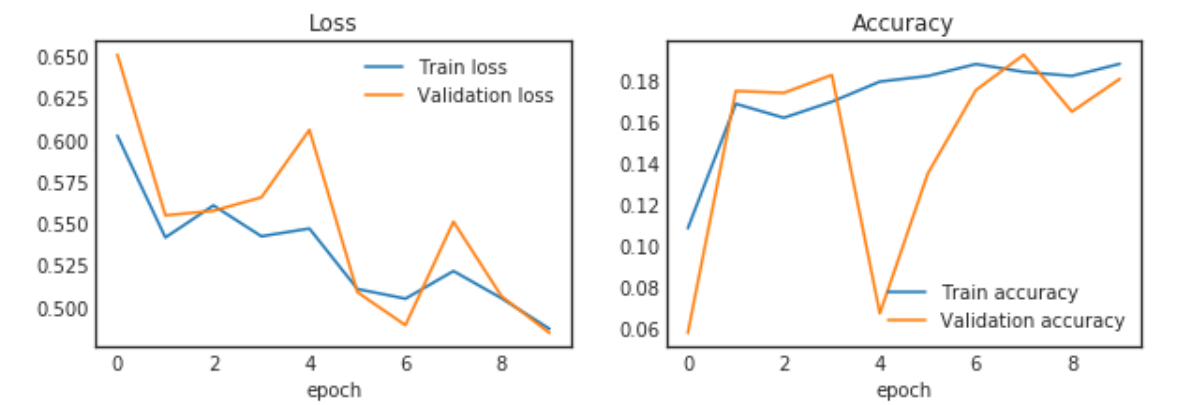
fig, (ax_loss, ax_acc) = plt.subplots(1, 2, figsize=(10,3))
ax_loss.plot(history.epoch, history.history["loss"], label="Train loss")
ax_loss.plot(history.epoch, history.history["val_loss"], label="Validation loss")
ax_acc.plot(history.epoch, history.history["acc"], label="Train accuracy")
ax_acc.plot(history.epoch, history.history["val_acc"], label="Validation accuracy")
plt.show()
Model3
df_depths = pd.read_csv('../input/depths.csv', index_col='id')
# df_depths.head()
df_depths.hist()
im_width = 128
im_height = 128
n_features = 1 # Number of extra features, like depth
path_train = '../input/train/'
train_ids = os.listdir(path_train+'images')
# Get and resize train images and masks
X = np.zeros((len(train_ids), im_height, im_width, 1), dtype=np.float32)
y = np.zeros((len(train_ids), im_height, im_width, 1), dtype=np.float32)
X_feat = np.zeros((len(train_ids), n_features), dtype=np.float32)
print('Getting and resizing train images and masks ... ')
# sys.stdout.flush()
for n, id_ in tqdm_notebook(enumerate(train_ids), total=len(train_ids)):
path = path_train
# Depth
X_feat[n] = df_depths.loc[id_.replace('.png', ''), 'z']
# Load X
img = load_img(path + '/images/' + id_, grayscale=True)
x_img = img_to_array(img)
x_img = resize(x_img, (128, 128, 1), mode='constant', preserve_range=True)
# Load Y
mask = img_to_array(load_img(path + '/masks/' + id_, grayscale=True))
mask = resize(mask, (128, 128, 1), mode='constant', preserve_range=True)
# Save images
X[n, ..., 0] = x_img.squeeze() / 255
y[n] = mask / 255
print('Done!')
X_train, X_valid, X_feat_train, X_feat_valid, y_train, y_valid = train_test_split(X, X_feat, y, test_size=0.15, random_state=42)
# Normalize X_feat
x_feat_mean = X_feat_train.mean(axis=0, keepdims=True)
x_feat_std = X_feat_train.std(axis=0, keepdims=True)
X_feat_train -= x_feat_mean
X_feat_train /= x_feat_std
X_feat_valid -= x_feat_mean
X_feat_valid /= x_feat_std
input_img = Input((im_width, im_hight, 1), name='img')
input_features = Input((n_features, ), name='feat')
c1 = Conv2D(8, (3, 3), activation='relu', padding='same') (input_img)
c1 = Conv2D(8, (3, 3), activation='relu', padding='same') (c1)
p1 = MaxPooling2D((2, 2)) (c1)
c2 = Conv2D(16, (3, 3), activation='relu', padding='same') (p1)
c2 = Conv2D(16, (3, 3), activation='relu', padding='same') (c2)
p2 = MaxPooling2D((2, 2)) (c2)
c3 = Conv2D(32, (3, 3), activation='relu', padding='same') (p2)
c3 = Conv2D(32, (3, 3), activation='relu', padding='same') (c3)
p3 = MaxPooling2D((2, 2)) (c3)
c4 = Conv2D(64, (3, 3), activation='relu', padding='same') (p3)
c4 = Conv2D(64, (3, 3), activation='relu', padding='same') (c4)
p4 = MaxPooling2D(pool_size=(2, 2)) (c4)
# Join features information in the depthest layer
f_repeat = RepeatVector(8*8)(input_features)
f_conv = Reshape((8, 8, n_features))(f_repeat)
p4_feat = concatenate([p4, f_conv], -1)
c5 = Conv2D(128, (3, 3), activation='relu', padding='same') (p4_feat)
c5 = Conv2D(128, (3, 3), activation='relu', padding='same') (c5)
u6 = Conv2DTranspose(64, (2, 2), strides=(2, 2), padding='same') (c5)
u6 = concatenate([u6, c4])
c6 = Conv2D(64, (3, 3), activation='relu', padding='same') (u6)
c6 = Conv2D(64, (3, 3), activation='relu', padding='same') (c6)
u7 = Conv2DTranspose(32, (2, 2), strides=(2, 2), padding='same') (c6)
u7 = concatenate([u7, c3])
c7 = Conv2D(32, (3, 3), activation='relu', padding='same') (u7)
c7 = Conv2D(32, (3, 3), activation='relu', padding='same') (c7)
u8 = Conv2DTranspose(16, (2, 2), strides=(2, 2), padding='same') (c7)
u8 = concatenate([u8, c2])
c8 = Conv2D(16, (3, 3), activation='relu', padding='same') (u8)
c8 = Conv2D(16, (3, 3), activation='relu', padding='same') (c8)
u9 = Conv2DTranspose(8, (2, 2), strides=(2, 2), padding='same') (c8)
u9 = concatenate([u9, c1], axis=3)
c9 = Conv2D(8, (3, 3), activation='relu', padding='same') (u9)
c9 = Conv2D(8, (3, 3), activation='relu', padding='same') (c9)
outputs = Conv2D(1, (1, 1), activation='sigmoid') (c9)
model3 = Model(inputs=[input_img, input_features], outputs=[outputs])
model3.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy']) #, metrics=[mean_iou]) # The mean_iou metrics seens to leak train and test values...
model3.summary()
epochs = 10
batch_size = 32
callbacks = [
EarlyStopping(patience=5, verbose=1),
ReduceLROnPlateau(patience=3, verbose=1)
]
results = model3.fit({'img': X_train, 'feat': X_feat_train}, y_train,
batch_size=batch_size, epochs=epochs, callbacks=callbacks,
validation_data=({'img': X_valid, 'feat': X_feat_valid}, y_valid),
verbose=1)
fig, (ax_loss, ax_acc) = plt.subplots(1, 2, figsize=(10,3))
ax_loss.plot(results.epoch, results.history["loss"], label="Train loss")
ax_loss.plot(results.epoch, results.history["val_loss"], label="Validation loss")
ax_loss.set_title('Loss')
ax_loss.set_xlabel('epoch')
ax_loss.legend(loc='best')
ax_acc.plot(results.epoch, results.history["acc"], label="Train accuracy")
ax_acc.plot(results.epoch, results.history["val_acc"], label="Validation accuracy")
ax_acc.set_title('Accuracy')
ax_acc.set_xlabel('epoch')
ax_acc.legend(loc='best')
plt.show()
Lovasz loss
# code download from: https://github.com/bermanmaxim/LovaszSoftmax
def lovasz_grad(gt_sorted):
"""
Computes gradient of the Lovasz extension w.r.t sorted errors
See Alg. 1 in paper
"""
gts = tf.reduce_sum(gt_sorted)
intersection = gts - tf.cumsum(gt_sorted)
union = gts + tf.cumsum(1. - gt_sorted)
jaccard = 1. - intersection / union
jaccard = tf.concat((jaccard[0:1], jaccard[1:] - jaccard[:-1]), 0)
return jaccard
# --------------------------- BINARY LOSSES ---------------------------
def lovasz_hinge(logits, labels, per_image=True, ignore=None):
"""
Binary Lovasz hinge loss
logits: [B, H, W] Variable, logits at each pixel (between -\infty and +\infty)
labels: [B, H, W] Tensor, binary ground truth masks (0 or 1)
per_image: compute the loss per image instead of per batch
ignore: void class id
"""
if per_image:
def treat_image(log_lab):
log, lab = log_lab
log, lab = tf.expand_dims(log, 0), tf.expand_dims(lab, 0)
log, lab = flatten_binary_scores(log, lab, ignore)
return lovasz_hinge_flat(log, lab)
losses = tf.map_fn(treat_image, (logits, labels), dtype=tf.float32)
loss = tf.reduce_mean(losses)
else:
loss = lovasz_hinge_flat(*flatten_binary_scores(logits, labels, ignore))
return loss
def lovasz_hinge_flat(logits, labels):
"""
Binary Lovasz hinge loss
logits: [P] Variable, logits at each prediction (between -\infty and +\infty)
labels: [P] Tensor, binary ground truth labels (0 or 1)
ignore: label to ignore
"""
def compute_loss():
labelsf = tf.cast(labels, logits.dtype)
signs = 2. * labelsf - 1.
errors = 1. - logits * tf.stop_gradient(signs)
errors_sorted, perm = tf.nn.top_k(errors, k=tf.shape(errors)[0], name="descending_sort")
gt_sorted = tf.gather(labelsf, perm)
grad = lovasz_grad(gt_sorted)
loss = tf.tensordot(tf.nn.relu(errors_sorted), tf.stop_gradient(grad), 1, name="loss_non_void")
return loss
# deal with the void prediction case (only void pixels)
loss = tf.cond(tf.equal(tf.shape(logits)[0], 0),
lambda: tf.reduce_sum(logits) * 0.,
compute_loss,
strict=True,
name="loss"
)
return loss
def flatten_binary_scores(scores, labels, ignore=None):
"""
Flattens predictions in the batch (binary case)
Remove labels equal to 'ignore'
"""
scores = tf.reshape(scores, (-1,))
labels = tf.reshape(labels, (-1,))
if ignore is None:
return scores, labels
valid = tf.not_equal(labels, ignore)
vscores = tf.boolean_mask(scores, valid, name='valid_scores')
vlabels = tf.boolean_mask(labels, valid, name='valid_labels')
return vscores, vlabels
def lovasz_loss(y_true, y_pred):
y_true, y_pred = K.cast(K.squeeze(y_true, -1), 'int32'), K.cast(K.squeeze(y_pred, -1), 'float32')
#logits = K.log(y_pred / (1. - y_pred))
logits = y_pred #Jiaxin
loss = lovasz_hinge(logits, y_true, per_image = True, ignore = None)
return loss
# model1 = load_model("./keras.model")
# remove last activation layer and use losvasz loss
input_x = model1.layers[0].input
output_layer = model1.layers[-1].input
model = Model(input_x, output_layer)
# c = optimizers.adam(lr = 0.01)
# lovasz_loss need input range (-∞,+∞), so cancel the last "sigmoid" activation
# Then the default threshod for pixel prediction is 0 instead of 0.5
model.compile(loss=lovasz_loss, optimizer='adam', metrics=['accuracy'])
early_stopping = EarlyStopping(patience=5, verbose=1)
# model_checkpoint = ModelCheckpoint("./keras.model", save_best_only=True, verbose=1)
reduce_lr = ReduceLROnPlateau(factor=0.1, patience=5, min_lr=0.00001, verbose=1)
epochs = 10
batch_size = 32
history = model.fit(x_train, y_train,
validation_data=[x_valid, y_valid],
epochs=epochs,
batch_size=batch_size,
#callbacks=[early_stopping, model_checkpoint, reduce_lr],
callbacks=[early_stopping, reduce_lr])
fig, (ax_loss, ax_acc) = plt.subplots(1, 2, figsize=(10,3))
ax_loss.plot(history.epoch, history.history["loss"], label="Train loss")
ax_loss.plot(history.epoch, history.history["val_loss"], label="Validation loss")
ax_loss.set_title('Loss')
ax_loss.set_xlabel('epoch')
ax_loss.legend(loc='best')
ax_acc.plot(history.epoch, history.history["acc"], label="Train accuracy")
ax_acc.plot(history.epoch, history.history["val_acc"], label="Validation accuracy")
ax_acc.set_title('Accuracy')
ax_acc.set_xlabel('epoch')
ax_acc.legend(loc='best')
plt.show()