Library
import numpy as np
from tqdm import tqdm
from sklearn.datasets import fetch_20newsgroups
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.utils.np_utils import to_categorical
from keras.layers import Dense, Input, Flatten
from keras.layers import Conv1D, MaxPooling1D, Embedding, Dropout
from keras.models import Model
Data
categories = ['alt.atheism', 'soc.religion.christian']
newsgroups_train = fetch_20newsgroups(subset='train', shuffle=True,
categories=categories,)
print (newsgroups_train.target_names)
print (len(newsgroups_train.data))
# print (newsgroups_train.data[1])
print("\n".join(newsgroups_train.data[0].split("\n")[10:15]))
%%time
texts = []
labels=newsgroups_train.target
texts = newsgroups_train.data
MAX_SEQUENCE_LENGTH = 1000
MAX_NB_WORDS = 20000
tokenizer = Tokenizer(num_words=MAX_NB_WORDS)
tokenizer.fit_on_texts(texts)
sequences = tokenizer.texts_to_sequences(texts)
print (sequences[0][:10])
word_index = tokenizer.word_index
print('Found %s unique tokens.' % len(word_index))
data = pad_sequences(sequences, maxlen=MAX_SEQUENCE_LENGTH)
print (data.shape)
print (data[0][200:250])
labels = to_categorical(np.array(labels))
print('Shape of data tensor:', data.shape)
print('Shape of label tensor:', labels.shape)
VALIDATION_SPLIT = 0.2
indices = np.arange(data.shape[0])
np.random.shuffle(indices)
data = data[indices]
labels = labels[indices]
nb_validation_samples = int(VALIDATION_SPLIT * data.shape[0])
x_train = data[:-nb_validation_samples]
y_train = labels[:-nb_validation_samples]
x_val = data[-nb_validation_samples:]
y_val = labels[-nb_validation_samples:]
print (x_train.shape)
print (y_train.shape)
print('Number of positive and negative reviews in traing and validation set ')
print (y_train.sum(axis=0))
print (y_val.sum(axis=0))
%%time
embeddings_index = {}
path = 'xxx'
f = open(path+'glove.6B.100d.txt')
for line in tqdm(f):
values = line.split(' ')
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
f.close()
print ()
print ('Found %s word vectors.' % len(embeddings_index))
EMBEDDING_DIM = 100
embedding_matrix = np.random.random((len(word_index) + 1, EMBEDDING_DIM))
for word, i in word_index.items():
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
# words not found in embedding index will be all-zeros.
embedding_matrix[i] = embedding_vector
print (embedding_matrix.shape)
print (embedding_matrix[0][:10])
Model 1
embedding_layer = Embedding(len(word_index) + 1, EMBEDDING_DIM,
weights=[embedding_matrix],
input_length=MAX_SEQUENCE_LENGTH,
trainable=False)
sequence_input = Input(shape=(MAX_SEQUENCE_LENGTH,), dtype='int32')
embedded_sequences = embedding_layer(sequence_input)
l_cov1= Conv1D(128, 5, activation='relu')(embedded_sequences)
l_pool1 = MaxPooling1D(5)(l_cov1)
l_cov2 = Conv1D(128, 5, activation='relu')(l_pool1)
l_pool2 = MaxPooling1D(5)(l_cov2)
l_cov3 = Conv1D(128, 5, activation='relu')(l_pool2)
l_pool3 = MaxPooling1D(35)(l_cov3) # global max pooling
l_flat = Flatten()(l_pool3)
l_dense = Dense(128, activation='relu')(l_flat)
preds = Dense(2, activation='softmax')(l_dense)
model = Model(sequence_input, preds)
model.summary()
model.compile(loss='categorical_crossentropy', optimizer='adam',metrics=['acc'])
history = model.fit(x_train, y_train, validation_data=(x_val, y_val),
epochs=10, batch_size=128)
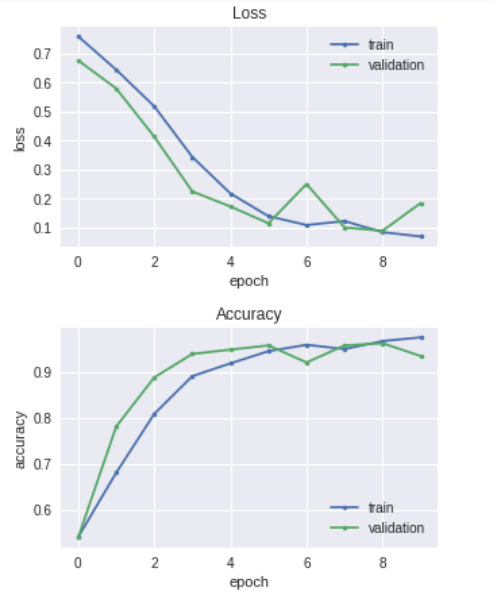
plt.figure(figsize =(5,3))
plt.plot(history.history['loss'], marker='.', label='train')
plt.plot(history.history['val_loss'], marker='.', label='validation')
plt.title('Loss')
plt.grid(True)
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(loc='best')
plt.show()
plt.figure(figsize =(5,3))
plt.plot(history.history['acc'], marker='.', label='train')
plt.plot(history.history['val_acc'], marker='.', label='validation')
plt.title('Accuracy')
plt.grid(True)
plt.xlabel('epoch')
plt.ylabel('accuracy')
plt.legend(loc='best')
plt.show()
Model 2
convs = []
filter_sizes = [3,4,5]
embedding_layer = Embedding(len(word_index) + 1, EMBEDDING_DIM,
weights=[embedding_matrix],
input_length=MAX_SEQUENCE_LENGTH,
trainable=True)
sequence_input = Input(shape=(MAX_SEQUENCE_LENGTH,), dtype='int32')
embedded_sequences = embedding_layer(sequence_input)
for fsz in filter_sizes:
l_conv = Conv1D(filters=128, kernel_size=fsz, activation='relu')(embedded_sequences)
l_pool = MaxPooling1D(5)(l_conv)
convs.append(l_pool)
l_merge = Concatenate(axis=1)(convs)
l_cov1= Conv1D(128, 5, activation='relu')(l_merge)
l_pool1 = MaxPooling1D(5)(l_cov1)
l_cov2 = Conv1D(128, 5, activation='relu')(l_pool1)
l_pool2 = MaxPooling1D(30)(l_cov2)
l_flat = Flatten()(l_pool2)
l_dense = Dense(128, activation='relu')(l_flat)
preds = Dense(2, activation='softmax')(l_dense)
model = Model(sequence_input, preds)
model.summary()
model.compile(loss='categorical_crossentropy', optimizer='adam',
metrics=['acc'])
history = model.fit(x_train, y_train, validation_data=(x_val, y_val),
epochs=10, batch_size=128)
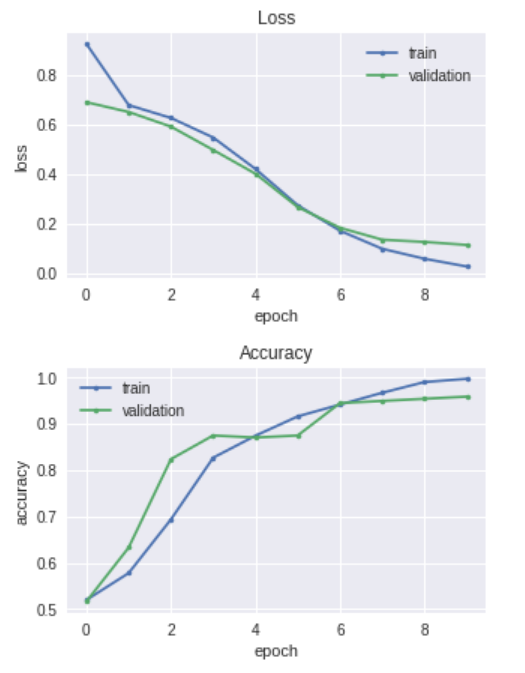
plt.figure(figsize =(5,3))
plt.plot(history.history['loss'], marker='.', label='train')
plt.plot(history.history['val_loss'], marker='.', label='validation')
plt.title('Loss')
plt.grid(True)
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(loc='best')
plt.show()
plt.figure(figsize =(5,3))
plt.plot(history.history['acc'], marker='.', label='train')
plt.plot(history.history['val_acc'], marker='.', label='validation')
plt.title('Accuracy')
plt.grid(True)
plt.xlabel('epoch')
plt.ylabel('accuracy')
plt.legend(loc='best')
plt.show()