動機
Average nucleotide identity (平均塩基一致率; ANI) をもとにゲノムをクラスタリングしたヒートマップが描きたい。
ペアワイズ (1vs1) の場合はANI calculatorがイケてるのだが、数が多くなるとちょっと大変。
そこで今回はANIを爆速で計算してくれるfastANIというソフトを使う。
今回はグラム陰性細菌Vibrio属のHarveyiクレードを例にとる。
ゲノムをダウンロードする
# Vibrio harveyi
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/770/115/GCF_000770115.1_ASM77011v2/GCF_000770115.1_ASM77011v2_genomic.fna.gz # ATCC_33843
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/009/665/315/GCF_009665315.1_ASM966531v1/GCF_009665315.1_ASM966531v1_genomic.fna.gz # 2011V-1164
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/908/435/GCF_001908435.2_ASM190843v2/GCF_001908435.2_ASM190843v2_genomic.fna.gz # QT520
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/009/184/745/GCF_009184745.1_ASM918474v1/GCF_009184745.1_ASM918474v1_genomic.fna.gz # WXL538
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/850/295/GCF_002850295.1_ASM285029v1/GCF_002850295.1_ASM285029v1_genomic.fna.gz # 345
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/558/435/GCF_001558435.2_ASM155843v2/GCF_001558435.2_ASM155843v2_genomic.fna.gz # FDAARGOS_107
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/591/145/GCF_001591145.1_ASM159114v1/GCF_001591145.1_ASM159114v1_genomic.fna.gz # NBRC_15634T
# Vibrio owensii
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/025/917/725/GCF_025917725.1_ASM2591772v1/GCF_025917725.1_ASM2591772v1_genomic.fna.gz # GL-605
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/887/655/GCF_002887655.1_ASM288765v1/GCF_002887655.1_ASM288765v1_genomic.fna.gz # 051011B
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/003/691/545/GCF_003691545.1_ASM369154v1/GCF_003691545.1_ASM369154v1_genomic.fna.gz # V180403
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/003/691/505/GCF_003691505.1_ASM369150v1/GCF_003691505.1_ASM369150v1_genomic.fna.gz # 1700302
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/310/575/GCF_001310575.2_ASM131057v2/GCF_001310575.2_ASM131057v2_genomic.fna.gz # SH14
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/021/755/GCF_002021755.1_ASM202175v1/GCF_002021755.1_ASM202175v1_genomic.fna.gz # XSBZ03
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/817/815/GCF_000817815.1_ASM81781v1/GCF_000817815.1_ASM81781v1_genomic.fna.gz # DY05T
# Vibrio campbellii
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/906/475/GCF_002906475.1_ASM290647v1/GCF_002906475.1_ASM290647v1_genomic.fna.gz # BoB-53
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/142/655/GCF_002142655.1_ASM214265v1/GCF_002142655.1_ASM214265v1_genomic.fna.gz # LA16-V1
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/022/453/845/GCF_022453845.1_ASM2245384v1/GCF_022453845.1_ASM2245384v1_genomic.fna.gz # IFL1
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/003/691/485/GCF_003691485.1_ASM369148v1/GCF_003691485.1_ASM369148v1_genomic.fna.gz # 170502
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/969/325/GCF_001969325.1_ASM196932v1/GCF_001969325.1_ASM196932v1_genomic.fna.gz # LMB29
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/024/442/195/GCF_024442195.1_ASM2444219v1/GCF_024442195.1_ASM2444219v1_genomic.fna.gz # LJC013
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/017/705/GCF_000017705.1_ASM1770v1/GCF_000017705.1_ASM1770v1_genomic.fna.gz # ATCC_BAA-1116T
gunzip *.gz
# Vibrio harveyi
mv GCF_000770115.1_ASM77011v2_genomic.fna ATCC_33843.fa
mv GCF_009665315.1_ASM966531v1_genomic.fna 2011V-1164.fa
mv GCF_001908435.2_ASM190843v2_genomic.fna QT520.fa
mv GCF_009184745.1_ASM918474v1_genomic.fna WXL538.fa
mv GCF_002850295.1_ASM285029v1_genomic.fna 345.fa
mv GCF_001558435.2_ASM155843v2_genomic.fna FDAARGOS_107.fa
mv GCF_001591145.1_ASM159114v1_genomic.fna NBRC_15634T.fa
# Vibrio owensii
mv GCF_025917725.1_ASM2591772v1_genomic.fna GL-605.fa
mv GCF_002887655.1_ASM288765v1_genomic.fna 051011B.fa
mv GCF_003691545.1_ASM369154v1_genomic.fna V180403.fa
mv GCF_003691505.1_ASM369150v1_genomic.fna 1700302.fa
mv GCF_001310575.2_ASM131057v2_genomic.fna SH14.fa
mv GCF_002021755.1_ASM202175v1_genomic.fna XSBZ03.fa
mv GCF_000817815.1_ASM81781v1_genomic.fna DY05T.fa
# Vibrio campbellii
mv GCF_002906475.1_ASM290647v1_genomic.fna BoB-53.fa
mv GCF_002142655.1_ASM214265v1_genomic.fna LA16-V1.fa
mv GCF_022453845.1_ASM2245384v1_genomic.fna IFL1.fa
mv GCF_003691485.1_ASM369148v1_genomic.fna 170502.fa
mv GCF_001969325.1_ASM196932v1_genomic.fna LMB29.fa
mv GCF_024442195.1_ASM2444219v1_genomic.fna LJC013.fa
mv GCF_000017705.1_ASM1770v1_genomic.fna ATCC_BAA-1116T.fa
fastANIを走らせる
ls -1|grep ".fa">in.list
fastANI --ql in.list --rl in.list -o fastani.out
less fastani.out
051011B.fa 051011B.fa 100 2079 2086
051011B.fa XSBZ03.fa 97.0118 1834 2086
051011B.fa GL-605.fa 96.9579 1874 2086
051011B.fa V180403.fa 96.9417 1864 2086
051011B.fa 1700302.fa 96.873 1852 2086
051011B.fa SH14.fa 96.8451 1851 2086
051011B.fa DY05T.fa 96.6277 1716 2086
051011B.fa LA16-V1.fa 91.3784 1621 2086
051011B.fa BoB-53.fa 91.3364 1557 2086
051011B.fa LMB29.fa 91.3347 1580 2086
051011B.fa IFL1.fa 91.327 1612 2086
051011B.fa 170502.fa 91.3196 1620 2086
051011B.fa ATCC_BAA-1116T.fa 90.7228 1497 2086
051011B.fa LJC013.fa 90.6381 1428 2086
051011B.fa 345.fa 90.2363 1520 2086
051011B.fa QT520.fa 90.1898 1611 2086
051011B.fa ATCC_33843.fa 90.1625 1597 2086
051011B.fa WXL538.fa 90.1469 1585 2086
051011B.fa FDAARGOS_107.fa 90.1217 1605 2086
051011B.fa 2011V-1164.fa 90.1025 1576 2086
051011B.fa NBRC_15634T.fa 90.0614 1490 2086
1700302.fa 1700302.fa 100 2132 2139
1700302.fa SH14.fa 99.8334 2054 2139
1700302.fa GL-605.fa 97.0249 1886 2139
1700302.fa V180403.fa 96.9478 1872 2139
1700302.fa XSBZ03.fa 96.9222 1807 2139
1700302.fa 051011B.fa 96.8728 1845 2139
1700302.fa DY05T.fa 96.6485 1717 2139
1700302.fa 170502.fa 91.4583 1623 2139
...
FASTAファイルに対応する菌種名を記入したtab-separated valueファイル(isolate.table)をつくる
isolate species
ATCC_33843.fa harveyi
2011V-1164.fa harveyi
QT520.fa harveyi
WXL538.fa harveyi
345.fa harveyi
FDAARGOS_107.fa harveyi
NBRC_15634T.fa harveyi
GL-605.fa owensii
051011B.fa owensii
V180403.fa owensii
1700302.fa owensii
SH14.fa owensii
XSBZ03.fa owensii
DY05T.fa owensii
BoB-53.fa campbellii
LA16-V1.fa campbellii
IFL1.fa campbellii
170502.fa campbellii
LMB29.fa campbellii
LJC013.fa campbellii
ATCC_BAA-1116T.fa campbellii
ヒートマップを描く
Rのpheatmapパッケージでヒートマップを描く。コードの参考サイト
# Set working directory
setwd("C:/path/to/directory")
library(dplyr)
library(reshape2)
library(pheatmap)
ani_tbl <- read.table("fastani.out") %>% select(V1, V2, V3)
isolate_metadata <- read.table("isolate.table", header = TRUE)
ani_mtrx <- acast(ani_tbl, V1~V2, value.var="V3")
distance = dist(ani_mtrx, method = "euclidean")
cluster = hclust(distance, method = "complete")
row.names(isolate_metadata) <- isolate_metadata$isolate
ani_matrix <- rownames(ani_mtrx)
ani_matrix <- isolate_metadata[isolate_metadata$isolate %in% ani_matrix,] %>% select(species)
ani_matrix <- ani_matrix %>% select(species)
my_colour = list(species = c("harveyi" = "#b56357", "owensii" = "#b4dbc0","campbellii"="#eae3ea"))
png("pheatmap.png", height=800, width=1000,res=120)
pheatmap(ani_mtrx, show_rownames = TRUE, cluster_rows = cluster, cluster_cols = cluster, annotation_row = ani_matrix, annotation_col = ani_matrix, annotation_colors = my_colour)
dev.off()
結果
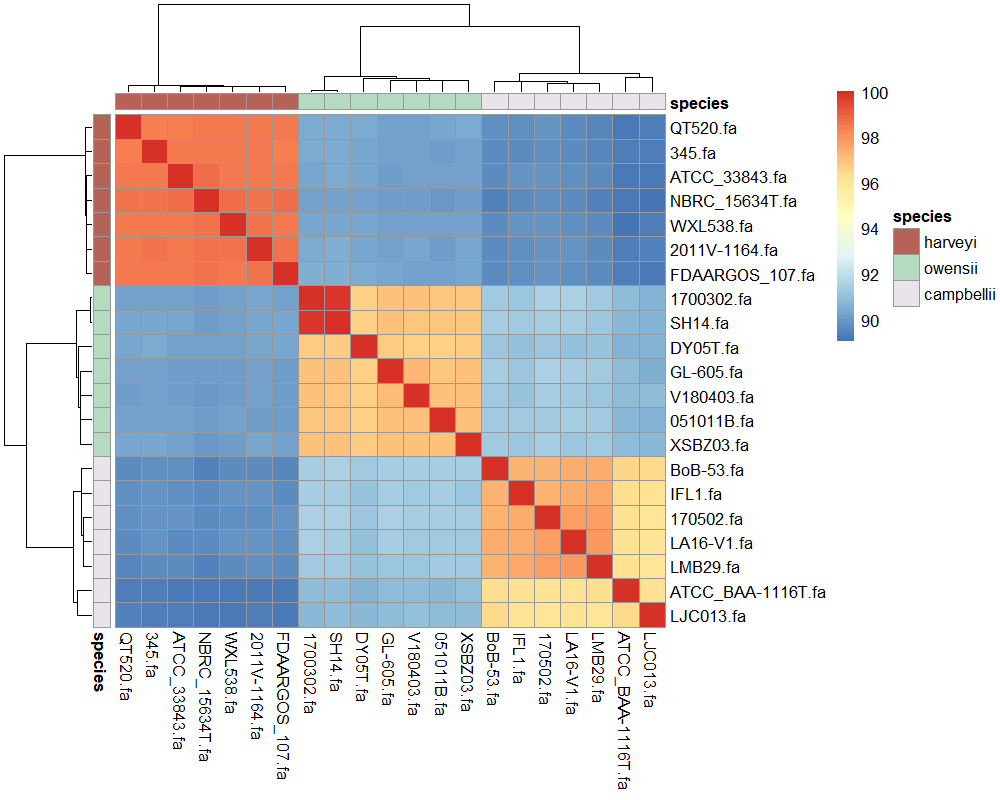
一般的に、ANIが95%以上で同一菌種とみなされる(それ未満で別種)。
今回の結果はそれをきれいに反映している。
経験上Harveyiクレードは16S rRNAの配列だと判別が難しいが、ゲノムレベルだときれいに分かれた。
参考サイト
https://github.com/ParBLiSS/FastANI
https://gitlab.mpi-bremen.de/jcifuent/Tonga_SOX/-/blob/master/scripts/fastANI_plot.Rmd