2025-05-23 追記:
gbdrawをリリースしました!
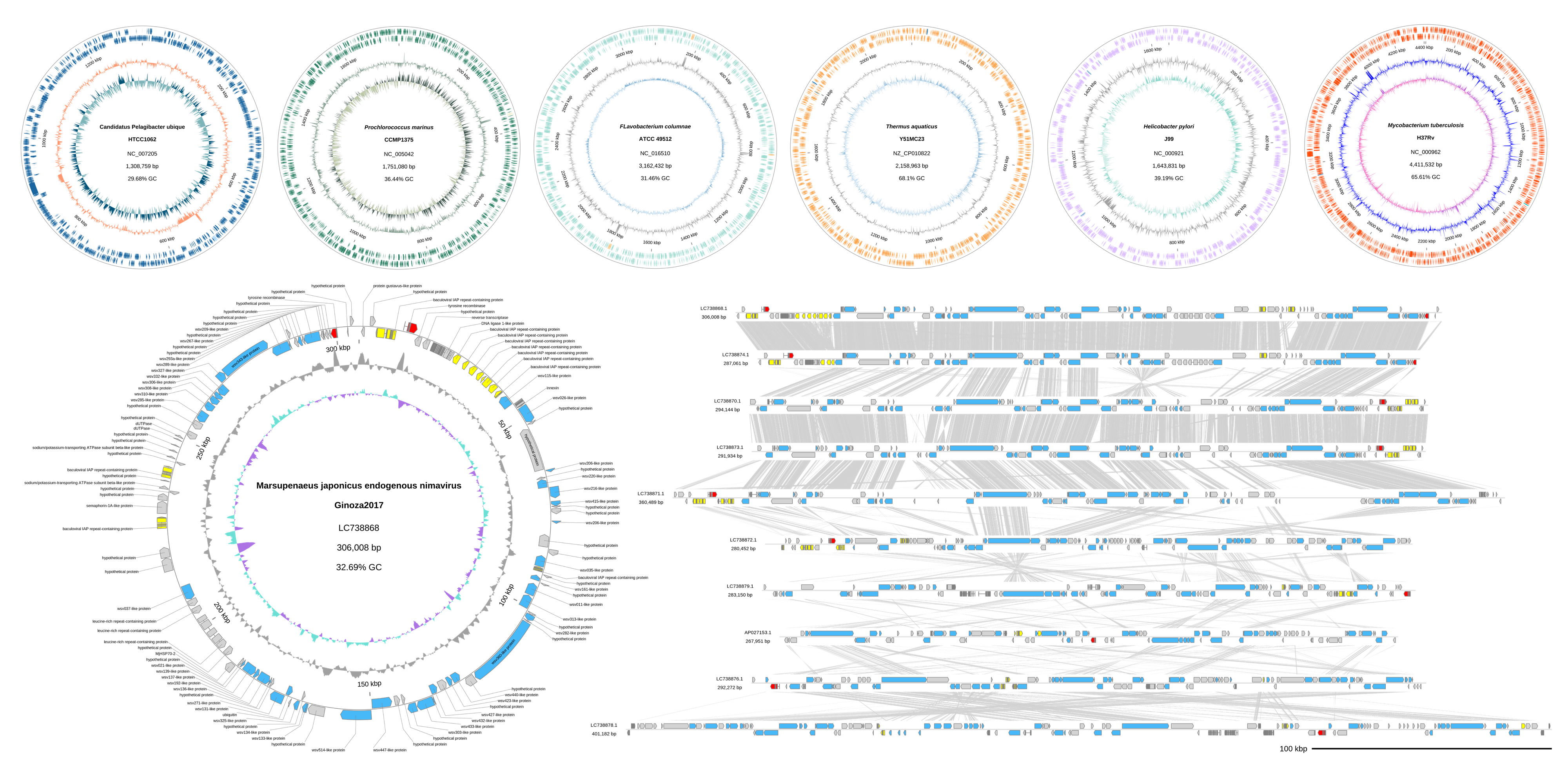
2024-02-06追記: 直鎖状・環状ゲノムを可視化するスクリプトをパッケージ化しています。お楽しみに
動機
ゲノムダイアグラム描画ソフトにはgggenomes、ClinkerやArtemis comparison tool (ACT)などがあるものの、PythonベースでSVG出力可能なものが欲しかった
plot_linear_genome.py
Generate plot in SVG
Requirements
Usage
$ ./plot_linear_genome.py
usage: plot_linear_genome.py [-h] -i [INPUT ...] [-b [BLAST ...]] [-t TABLE] [-o OUTPUT] [-n NT] [-w WINDOW] [-s STEP]
[--separate_strands] [--show_gc] [--align_center] [--evalue EVALUE] [--bitscore BITSCORE]
[--identity IDENTITY]
Generate plot in SVG
optional arguments:
-h, --help show this help message and exit
-i [INPUT ...], --input [INPUT ...]
genbank (required)
-b [BLAST ...], --blast [BLAST ...]
input BLAST result file in tab-separated format (-outfmt 6 or 7) (optional)
-t TABLE, --table TABLE
color table (optional)
-o OUTPUT, --output OUTPUT
output prefix (default: diagram)
-n NT, --nt NT dinucleotide (default: GC).
-w WINDOW, --window WINDOW
window size (default: 1000)
-s STEP, --step STEP step size (default: 100)
--separate_strands separate forward and reverse strands (default: False). Features of undefined strands are shown
on the forward strand.
--show_gc plot GC content below genome (default: False).
--align_center Align genomes to the center (default: False).
--evalue EVALUE evalue threshold (default=1e-2)
--bitscore BITSCORE bitscore threshold (default=50)
--identity IDENTITY identity threshold (default=0)
GenBank形式ファイル(.gb or .gbk)とBLASTN/TBLASTX出力ファイル(任意; -outfmt 6 or 7)を入力する。
BLAST出力ファイルは、GenBank形式ファイルと同じ配列かつIDのFASTAファイルをもとに作成する。
例としてリンホシスチス病ウイルスのゲノムダイアグラムを描く。
- Lymphocystis disease virus 1, complete genome (NC_001824.1)
- Lymphocystis disease virus Sa isolate SA9, complete genome (NC_033423.1)
- Lymphocystis disease virus 2 LCDV-JP_Oita_2018 DNA, complete genome (LC534415.1)
FASTA形式のゲノム配列ファイルについてTBLASTX (or BLASTN; -outfmt 7)で相同性検索
$ tblastx -query NC_001824.fasta -subject NC_033423.fasta -outfmt 7 -out NC_001824_NC_033423.tblastx.out
$ tblastx -query NC_033423.fasta -subject LC534415.fasta -outfmt 7 -out NC_033423_LC534415.tblastx.out
ゲノムダイアグラムを描く。ファイルの順番に注意されたい。
./plot_linear_genome.py -i NC_001824.gb NC_033423.gb LC534415.gb -b NC_001824_NC_033423.tblastx.out NC_033423_LC534415.tblastx.out --separate_strands --align_center --evalue 1e-10 --bitscore 100 # output: out.svg
