Background
いろいろな判別分析を比較してみた「Rで判別分析いろいろ(11種類+ Deep Learning)」 とか 「眼鏡っ娘分類システムの改良(判別分析12種類:SVM, Random Forest, Deep Learning 他)」 が、毎回チューニングするのに結構時間がかかる。主な原因は3つ
- それぞれ違ったパッケージを使っていたので、設定方法が違ったりして調べるのに時間がかかる
- グリッドサーチが実装されていない場合は自力実装
- 並列化とか結構面倒
そこで、すこし前に見つけて、いつかやってみようとおもっていた「caret」
- いろいろなパッケージのwapperになっていて、同じ使い勝手でグリッドサーチができるようになっている
- 並列化も勝手にやってくれる。とてもカンタン
- すごくたくさんの手法が使える
Summary
方法
- データ・セットは2値分類で、お手軽につかえるkernlabパッケージのspam
- トレーニング用とテスト用にデータを分けておく
- いろいろな分類手法を、caretでトレーニング&自動的にチューニングする
- テストデータで正解率を計算
- 各モデルの正解率を比較する
結果
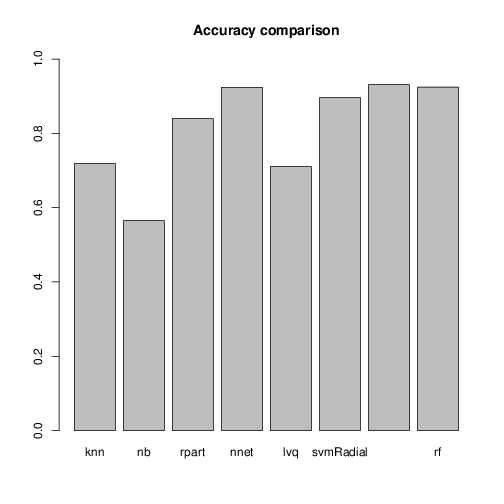
| 手法 | caretで指定するmethod名 | 正解率 |
|---|---|---|
| k Nearest Neighbor | knn | 0.725 |
| naive Bayse | nb | 0.565 |
| Decision Tree | rpart | 0.840 |
| 3層ニューラルネット | nnet | 0.921 |
| 学習ベクトル量子化ニューラルネットワーク | lvq | 0.706 |
| Support Vector Machine | svmRadial | 0.900 |
| Gradient Boosting Machine | gbm | 0.930 |
| Random Forest | rf | 0.920 |
結論
- チューニング済のモデルで正解率を計算した。3層ニューラルネット、Gradient Boosting Model, Random Forest、次いでSVMが良かった。
- 個別のパッケージを使うよりも、コードが圧倒的に見やすい。グリッドサーチを自分で書かなくてよい。自分で並列化しなくてよい。
- チューニング結果も表示可能でわかりやすい
つまり、すばらしい、ということ。
感想
簡単すぎてびっくり。
チューニング方法は、全自動だけではなく、細かく設定できるので、もっと使い込んでいきたい。
References...1つだけ読むなら
英語大丈夫な人は断然ここ
http://topepo.github.io/caret/training.html
英語嫌いな人は、ここがおすすめ
http://qiita.com/aokikenichi/items/46ba5e5d15927ac12c5f
Details
使用したデータ
- お手軽につかえるkernlabパッケージのspam
- 4601通のメールをspamとnon-spamに分類してあるデータ(57次元)
- 460通を学習データ、残りを検証データに使った(以前とは逆にした)
Source code
8種類のチューニングがシンプルに仕上がった。
#
# 判別分析いろいろ on caret
#
# openning
starttime <- proc.time() # 処理時間計測のため
# ライブラリの準備
library(caret) # caret
library(dplyr) #
library(doMC) # 並列計算
registerDoMC(cores = detectCores()) # 使用するコア数を決定。
# dataset
library(kernlab)
data(spam)
dim(spam)
data<-spam
num<-10*(1:(nrow(data)/10)) # 10行おきにサンプル作成
data.test<-data[-num,]
data.train<-data[num,]
#### methodを引数とする関数にして、汎用化する ####
predictionMethod <- function(methodname, traindata, testdata){
# model作成
m <- train(
data = traindata,
type ~ .,
method = methodname
)
# テストデータで予測
p <- predict(m, newdata = testdata)
# accuracy
t<-table(testdata[,ncol(testdata)],p) # confusion tableの作成
a<-sum(t[row(t)==col(t)])/sum(t) # accuracyの計算
# 戻り値は、リスト(メソッド名、予測結果、正解率、confusion table、モデル)
return(list(methodname,p,a,t,m))
}
# 比較
method_list <- c("knn","nb","rpart","nnet","lvq","svmRadial","gbm","rf")
accuracies <- vector()
for(i in method_list){
r <- predictionMethod(i, data.train, data.test)
# 結果を出力
plot(r[[2]],main=r[[1]]) # 予測結果(分類結果) titleにmethod名
# なぜか一度変数に代入したからprintしないとうまく描画されない
# カッコ悪いがこのままつづける
p_model<-plot(r[[5]],main=r[[1]]) # Model titleにmethod名
print(p_model)
print(r[[1]]) # method
print(r[[3]]) # accuracy
print(r[[4]]) # confusion table
accuracies <- c(accuracies, r[[3]])
}
# 各手法の成績を比較
barplot(accuracies, names.arg = method_list, ylim = 0:1, main="Accuracy comparison")
# closing
proc.time()-starttime # 処理時間計算し表示
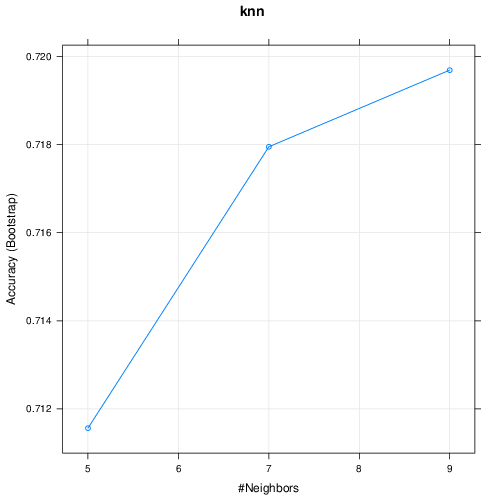
k Nearest Neighbor (knn)
k-Nearest Neighbors
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
k Accuracy Kappa
5 0.7199522 0.4145264
7 0.7178108 0.4094426
9 0.7096166 0.3900999
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was k = 5.
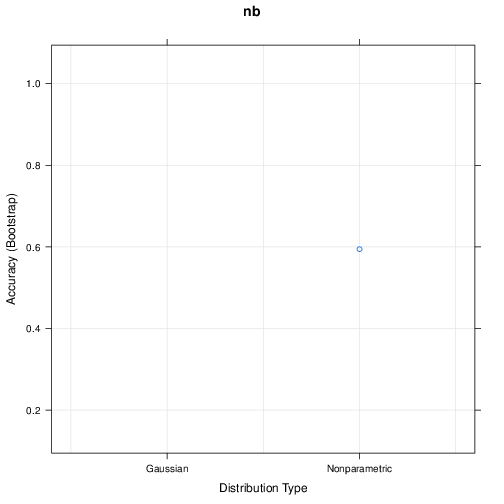
naive Bayse (nb)
Naive Bayes
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
usekernel Accuracy Kappa
FALSE NaN NaN
TRUE 0.6028973 0.2952961
Tuning parameter 'fL' was held constant at a value of 0
Tuning
parameter 'adjust' was held constant at a value of 1
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were fL = 0, usekernel = TRUE and adjust
= 1.
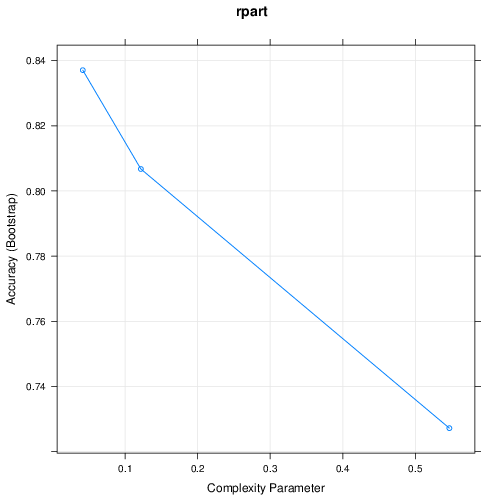
Decision Tree (rpart)
CART
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
cp Accuracy Kappa
0.04143646 0.8465669 0.6764833
0.12154696 0.8171015 0.6026779
0.54696133 0.7282228 0.3510546
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was cp = 0.04143646.
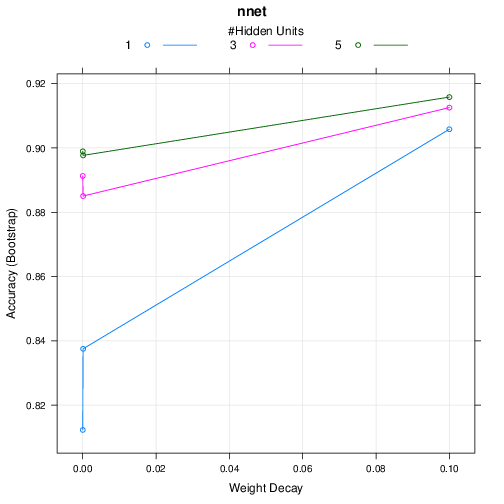
3層ニューラルネット (nnet)
Neural Network
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
size decay Accuracy Kappa
1 0e+00 0.8216229 0.6265190
1 1e-04 0.8649766 0.7150029
1 1e-01 0.8943997 0.7806125
3 0e+00 0.8867312 0.7656458
3 1e-04 0.8924993 0.7754566
3 1e-01 0.9068637 0.8042872
5 0e+00 0.8854677 0.7630163
5 1e-04 0.8927262 0.7746208
5 1e-01 0.9099463 0.8107649
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were size = 5 and decay = 0.1.
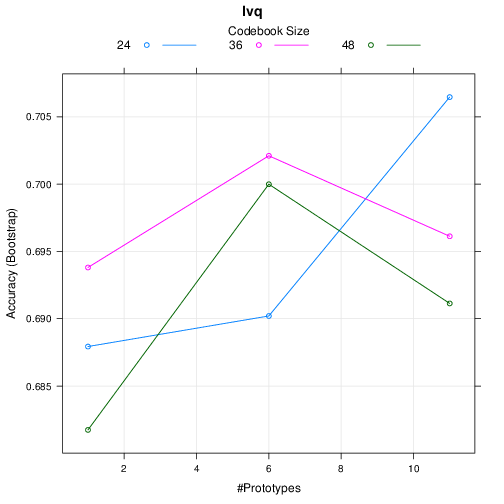
学習ベクトル量子化ニューラルネットワーク (lvq)
Learning Vector Quantization
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
size k Accuracy Kappa
24 1 0.6857730 0.3347985
24 6 0.7062956 0.3806374
24 11 0.7030803 0.3728038
36 1 0.7050989 0.3734752
36 6 0.7027073 0.3687149
36 11 0.7073002 0.3852150
48 1 0.7137609 0.3967521
48 6 0.7075282 0.3769616
48 11 0.7139488 0.3970061
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were size = 48 and k = 11.
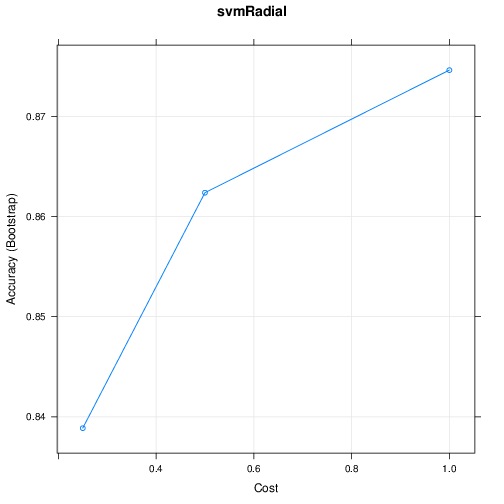
Support Vector Machine (svmRadial)
Support Vector Machines with Radial Basis Function Kernel
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
C Accuracy Kappa
0.25 0.8538915 0.6822898
0.50 0.8787777 0.7402555
1.00 0.8877831 0.7611116
Tuning parameter 'sigma' was held constant at a value of 0.02496736
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were sigma = 0.02496736 and C = 1.
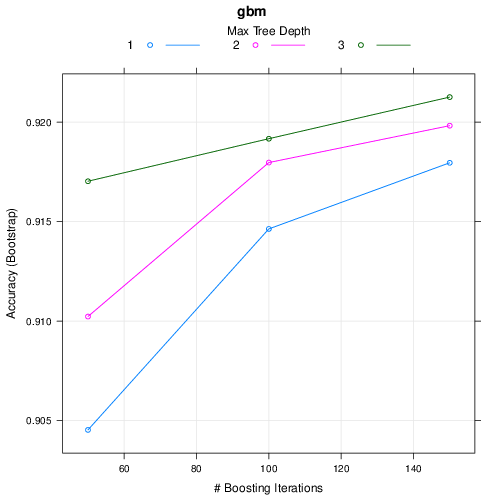
Gradient Boosting Machine (gbm)
Stochastic Gradient Boosting
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
interaction.depth n.trees Accuracy Kappa
1 50 0.9091337 0.8046674
1 100 0.9233847 0.8362842
1 150 0.9238490 0.8373352
2 50 0.9160698 0.8203036
2 100 0.9249554 0.8394724
2 150 0.9266512 0.8436142
3 50 0.9219410 0.8330738
3 100 0.9262219 0.8430003
3 150 0.9287916 0.8483900
Tuning parameter 'shrinkage' was held constant at a value of 0.1
Tuning parameter 'n.minobsinnode' was held constant at a value of 10
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were n.trees = 150, interaction.depth =
3, shrinkage = 0.1 and n.minobsinnode = 10.
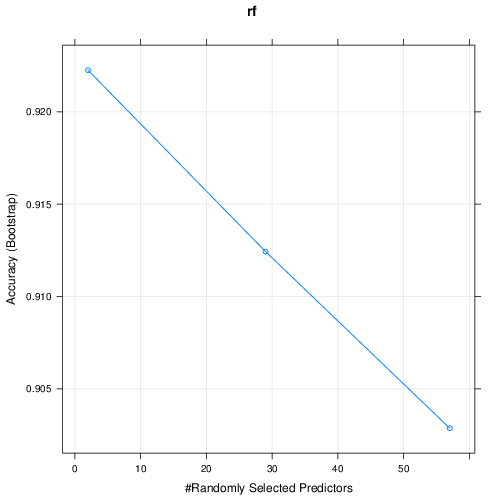
Random Forest (rf)
Random Forest
460 samples
57 predictor
2 classes: 'nonspam', 'spam'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 460, 460, 460, 460, 460, 460, ...
Resampling results across tuning parameters:
mtry Accuracy Kappa
2 0.9275362 0.8450614
29 0.9169599 0.8237313
57 0.9038631 0.7961136
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mtry = 2.
References
- http://topepo.github.io/caret/
- http://topepo.github.io/caret/training.html
- http://topepo.github.io/caret/modelList.html
- http://d.hatena.ne.jp/isseing333/20101022/1287735709
- http://kanosuke.hatenadiary.jp/entry/2015/10/19/231724
- http://tjo.hatenablog.com/entry/2013/09/02/190449
- http://stackoverflow.com/questions/22275173/creategrid-function-from-caret-package-was-it-removed
- http://testblog234wfhb.blogspot.jp/2014/06/random-forest-tuning-by-caret.html
- https://cran.r-project.org/web/packages/caret/index.html
- http://qiita.com/aokikenichi/items/46ba5e5d15927ac12c5f