Lorentz(1963)の方程式
カオスの研究で有名な方程式です。
\frac{\partial x_1}{\partial t} = \sigma (x_2 - x_1)
\frac{\partial x_2}{\partial t} = x_1(\rho-x_3) -x_2
\frac{\partial x_3}{\partial t} = x_1 x_2 - \beta x_3
以下の条件で解きます。パラメータ
\sigma = 10, \rho=28, \beta = 8/3
初期値
(x_1, x_2, x_3)=(1, 0, 0)
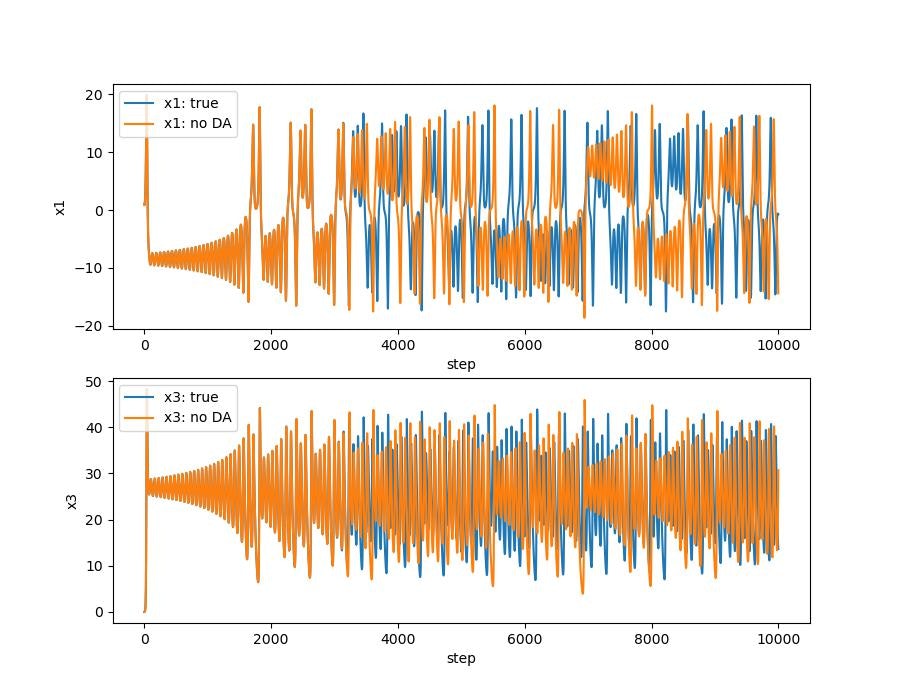
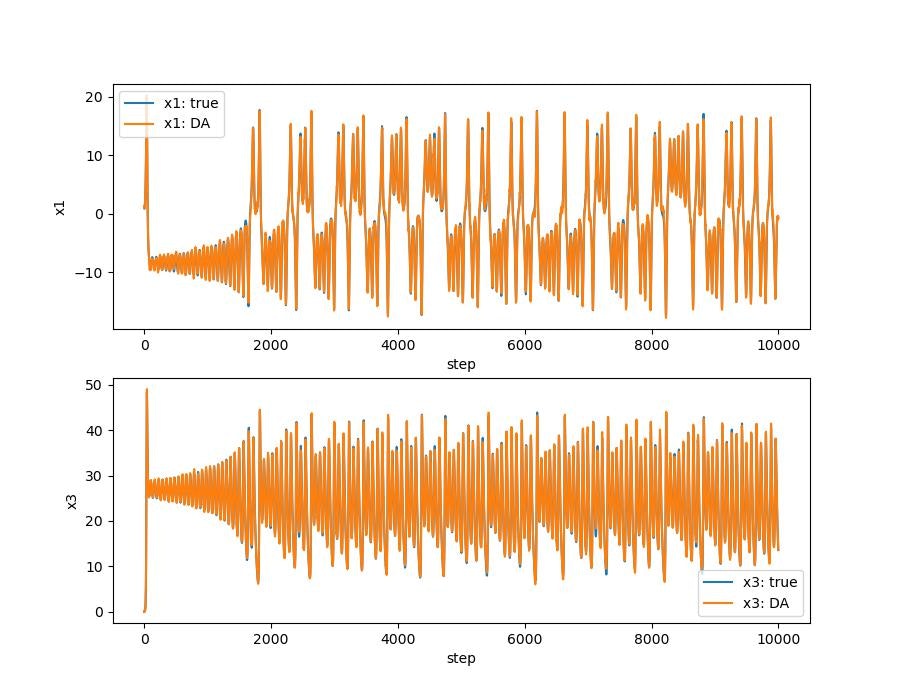
データ同化ありなしの比較
ローレンツ・アトラクタ。はじめのうちは3本とも重なっていますが,途中からずれてます。データ同化する(オレンジ)と真値とほぼ同様の解になります。
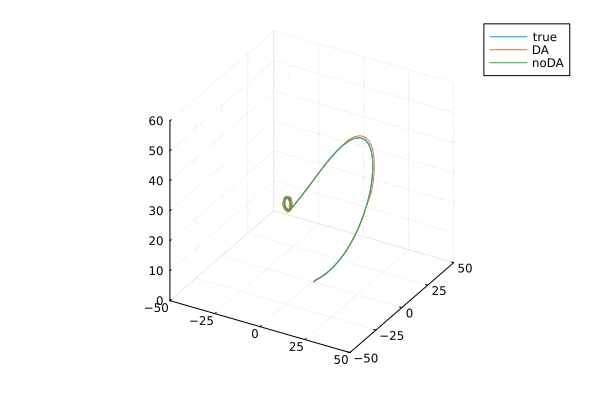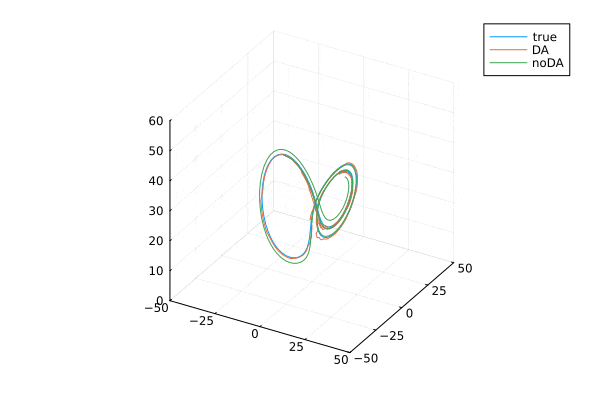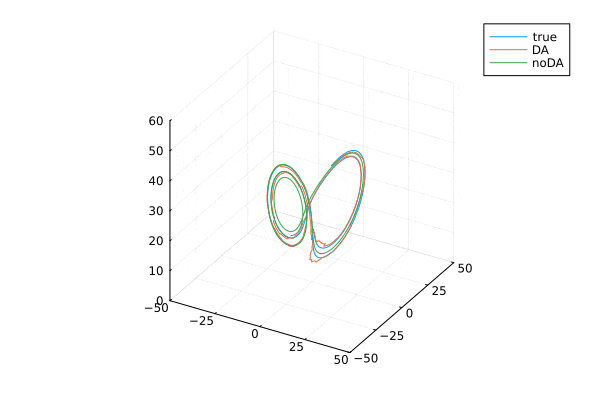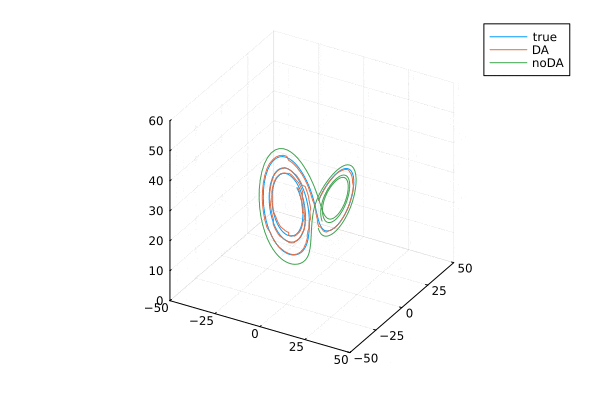
データ同化
Lorentz96の方は下記。
今回の数値解法は上記と同様,4次精度のルンゲ=クッタです。
using PyPlot
using Random
using LinearAlgebra
using HDF5
function Lorentz63(x::Vector{Float64})
σ::Float64 = 10
ρ::Float64 = 28
β::Float64 = 8/3
Lorentz63 = zeros(3)
Lorentz63[1] = σ*(x[2]-x[1])
Lorentz63[2] = x[1]*(ρ - x[3]) - x[2]
Lorentz63[3] = x[1]*x[2] - β*x[3]
return Lorentz63
end
function RK4(f, dt, x)
k1 = f(x)
k2 = f(x + k1*dt*0.5)
k3 = f(x + k2*dt*0.5)
k4 = f(x + k3*dt)
return x + dt*(k1 + 2k2 + 2k3 + k4)/6.
end
# 観測値の生成
function makeobs(y, xtrue, dstep, nmax)
Random.seed!(10)
for i=1:nmax
if mod(i, dstep) == 0
y[:,i] .= xtrue[:,i] .+ randn()
end
end
end
# Jacobianの生成
function makeM(x, N, dt)
delta = 1e-2
E = Matrix{Float64}(I, N, N)
M = zeros(Float64, N, N)
for j=1:N
M[:, j] = (RK4(Lorentz63, dt, x + delta*E[:, j]) - RK4(Lorentz63, dt, x))/delta
end
return M
end
function main()
nmax = 10000
dstep = 5
dt = 0.01
x0 = zeros(3, nmax+1)
x1 = zeros(3, nmax+1)
x0[:,1] = [1.0, 0., 0.]
x1[:,1] = [1.00001, 0., 0.]
# x0が真値,x1が真値と初期値が若干異なる解
for i=1:nmax
x0[:, i+1] = RK4(Lorentz63, dt, x0[:,i])
x1[:, i+1] = RK4(Lorentz63, dt, x1[:,i])
end
#println(x)
y = zeros(3, nmax +1)
makeobs(y, x0, dstep, nmax)
N = 3
Xa = zeros(Float64, N, nmax+1)
Xf = zeros(Float64, N, nmax+1)
Pa = zeros(Float64, N, N, nmax+1)
Pf = zeros(Float64, N, N, nmax+1)
R = Matrix{Float64}(I, N, N)
K = zeros(Float64, N, N)
H = Matrix{Float64}(I, N, N)
α = 1.5 # inflation factor
println(Xa[:,1])
Pa[:,:,1] = Matrix{Float64}(20I, N, N)
Xa[:,1] = [1.00001, 0., 0.]
println(Xa[:,1])
for i=1:nmax
Xf[:, i+1] = RK4(Lorentz63, dt, Xa[:,i])
if mod(i+1, dstep) == 0
M = makeM(Xf[:,i+1], N, dt)
Pf[:,:,i+1] = α*M*Pa[:,:,i]*M'
K = Pf[:,:,i+1]*H'*inv(H*Pf[:,:,i+1]*H' + R)
Xa[:,i+1] = Xf[:,i+1] + K*(y[:,i+1]-H*Xf[:,i+1])
Pa[:,:,i+1] = (I - K*H)*Pf[:,:,i+1]
else
Xa[:,i+1] = Xf[:,i+1]
Pa[:,:, i+1] = Pa[:,:, i]
end
end
# 結果の可視化
plot_start=1
plot_end=10000
fig = plt.figure(figsize=(9, 7.))
ax1 = fig.add_subplot(211)
ax1.plot(x0[1,plot_start:plot_end], label="x1: true")
ax1.plot(x1[1,plot_start:plot_end], label="x1: no DA")
#ax1.plot(y[1,plot_start:plot_end], label="x1: obs.", "o", markersize=2.5)
ax2 = fig.add_subplot(212)
ax2.plot(x0[3,plot_start:plot_end], label="x3: true")
ax2.plot(x1[3,plot_start:plot_end], label="x3: no DA")
#ax2.plot(y[3,plot_start:plot_end], label="x3: obs.", "o", markersize=2.5)
ax1.set_xlabel("step")
ax1.set_ylabel("x1")
ax1.legend()
ax2.set_xlabel("step")
ax2.set_ylabel("x3")
ax2.legend()
plt.savefig("true_noDA.jpeg")
fig = plt.figure(figsize=(9, 7.))
ax1 = fig.add_subplot(211)
ax1.plot(x0[1,plot_start:plot_end], label="x1: true")
ax1.plot(Xa[1,plot_start:plot_end], label="x1: DA",)
#ax1.plot(y[1,plot_start:plot_end], label="u1", "o", markersize=2.5)
ax2 = fig.add_subplot(212)
ax2.plot(x0[3,plot_start:plot_end], label="x3: true")
ax2.plot(Xa[3,plot_start:plot_end], label="x3: DA",)
#ax2.plot(y[3,plot_start:plot_end], label="u3", "o", markersize=2.5)
ax1.set_xlabel("step")
ax1.set_ylabel("x1")
ax1.legend()
ax2.set_xlabel("step")
ax2.set_ylabel("x3")
ax2.legend()
plt.savefig("true_DA.jpeg")
rms_a = zeros(nmax+1)
rms_o = zeros(nmax+1)
trPa = zeros(nmax+1)
for i=1:nmax
trPa[i] = sqrt(tr(Pa[:,:,i])/N)
rms_a[i] = norm(Xa[:,i]-x0[:,i])/sqrt(N)
rms_o[i] = norm(y[:,i]-x0[:,i])/sqrt(N)
end
fig = plt.figure()
ax1 = fig.add_subplot(111)
ax1.set_ylim(-1,10)
ax1.plot(rms_a[plot_start:plot_end], label="ana.")
ax1.plot(rms_o[plot_start:plot_end], label="obs.", "o", markersize=2.5)
ax1.plot(trPa[plot_start:plot_end], label="Trace")
ax1.legend()
ax = plt.figure().add_subplot(projection="3d")
ax.plot(x0[1,plot_start:plot_end], x0[2,plot_start:plot_end], x0[3,plot_start:plot_end], )
ax.plot(Xa[1,plot_start:plot_end], Xa[2,plot_start:plot_end], Xa[3,plot_start:plot_end], )
#ax.plot(x1[1,100:300], x1[2,100:300], x1[3,100:300], )
return x0, x1, Xa
end
実行と真値とデータ同化結果,データ同化なしの結果の出力
xtrue, x_noDA, Xa = main()
h5open("L63_true_da.h5", "w") do file
write(file, "xtrue", xtrue)
write(file, "Xa", Xa)
write(file, "x_noDA", x_noDA)
end
ローレンツアトラクタの可視化
using Plots
using HDF5
file = h5open("L63_true_da.h5", "r")
xtrue = read(file, "xtrue")
x_noDA = read(file, "x_noDA")
Xa = read(file, "Xa")
anim = @animate for i=2:100:9000
plot(xtrue[1,i:i+300], xtrue[2,i:i+300],xtrue[3,i:i+300], label="true",
xlim = (-50, 50), ylim = (-50, 50), zlim = (0, 60),)
plot!(Xa[1,i:i+300], Xa[2,i:i+300], Xa[3,i:i+300], label="DA",
xlim = (-50, 50), ylim = (-50, 50), zlim = (0, 60),)
plot!(x_noDA[1,i:i+300], x_noDA[2,i:i+300], x_noDA[3,i:i+300], label="noDA",
xlim = (-50, 50), ylim = (-50, 50), zlim = (0, 60),)
#plot!(sol2[1,i:i+300], sol2[2,i:i+300], sol2[3,i:i+300],
# xlim = (-50, 50), ylim = (-50, 50), zlim = (0, 60), label="u0_2")
end
gif(anim, "lorentz3D-da.gif", fps = 2)