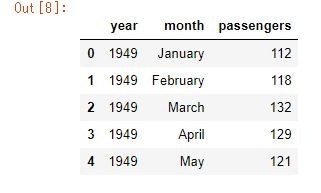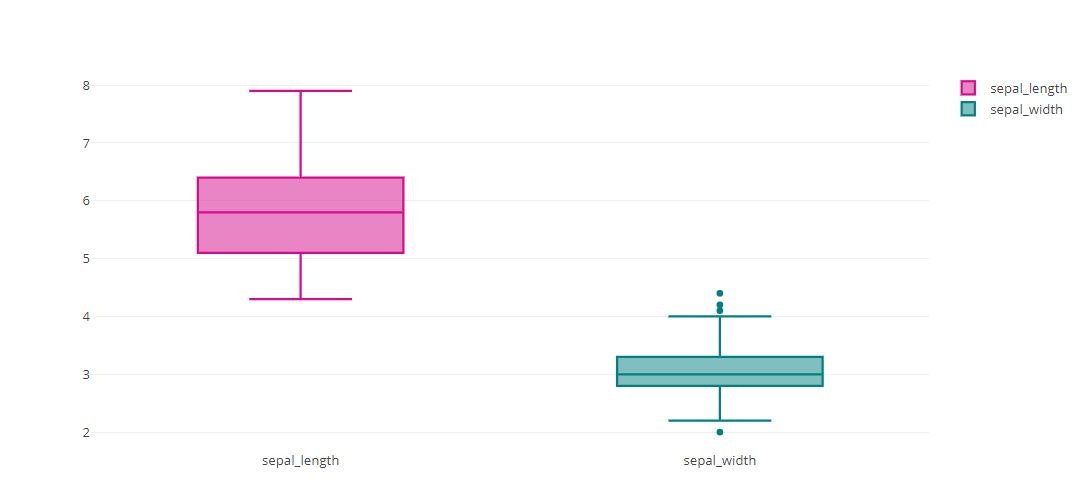はじめに
スーパーでブロッコリーを見るとPlotlyを連想する病気にかかっています。
自分の作業用によく使うPlotlyの使い方をまとめたので公開します。
実際にグリグリ動かしたい方はkaggleのkernelにアップしてるのでそちらでグリグリしてください。
https://www.kaggle.com/kageyama/how-to-use-plotly
※ちなみにPlotlyの3Dグラフをグリグリしたことがない人は、是非とも一度グリグリした方がいい。感動する。
データロード
データはseabornから
iris,titanic,flightsの3つを適宜使用する。
import numpy as np
import pandas as pd
import plotly as py
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.figure_factory as ff
import seaborn as sns
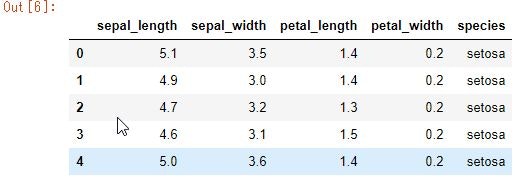
iris_data = sns.load_dataset("iris")
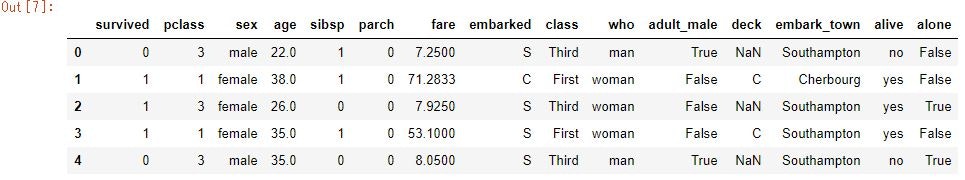
titanic_data = sns.load_dataset('titanic')
flights_data = sns.load_dataset('flights')
iris_data.head()
titanic_data.head()
flights_data.head()
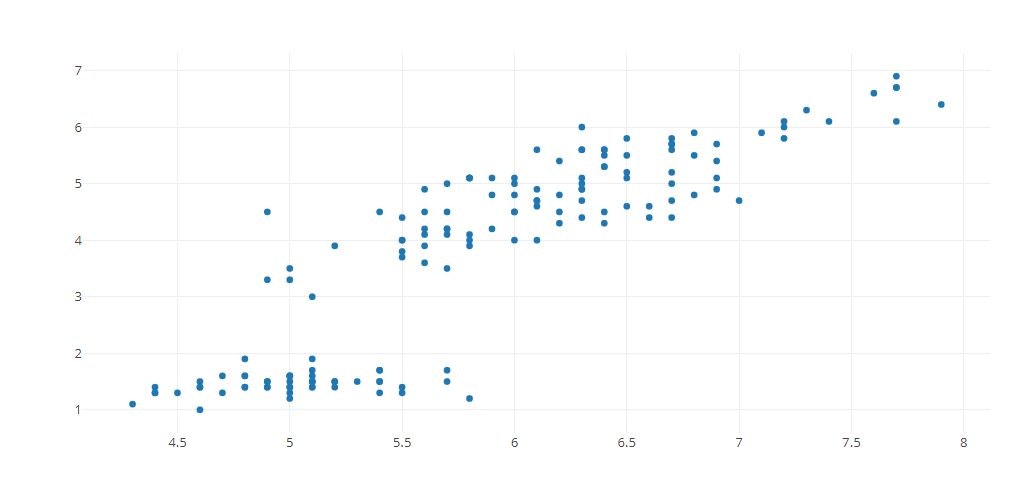
Scatter Plot
# Create a trace
trace = go.Scatter(
x = iris_data['sepal_length'] ,
y = iris_data['petal_length'] ,
mode = 'markers'
)
tmp = [trace]
# Plot and embed in ipython notebook!
py.offline.iplot(tmp)
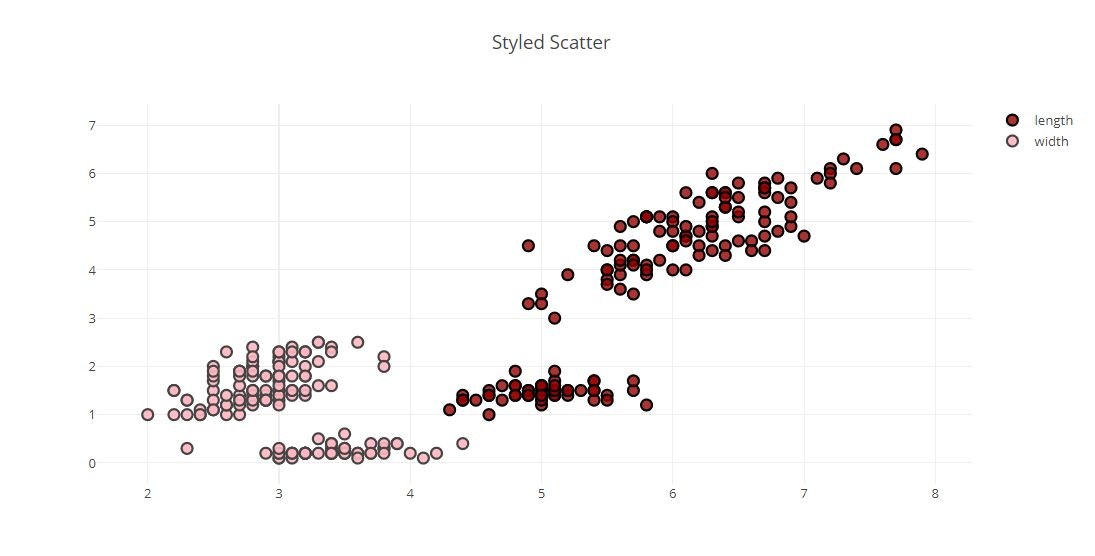
# Style Scatter Plots
trace0 = go.Scatter(
x = iris_data['sepal_length'],
y = iris_data['petal_length'],
name = 'length',
mode = 'markers',
marker = dict(
size = 10,
color = 'rgba(152, 0, 0, .8)',
line = dict(
width = 2,
color = 'rgb(0, 0, 0)'
)
)
)
trace1 = go.Scatter(
x = iris_data['sepal_width'],
y = iris_data['petal_width'],
name = 'width',
mode = 'markers',
marker = dict(
size = 10,
color = 'rgba(255, 182, 193, .9)',
line = dict(
width = 2,
)
)
)
tmp = [trace0, trace1]
layout = dict(title = 'Styled Scatter',
yaxis = dict(zeroline = False),
xaxis = dict(zeroline = False)
)
fig = dict(data=tmp, layout=layout)
py.offline.iplot(fig)
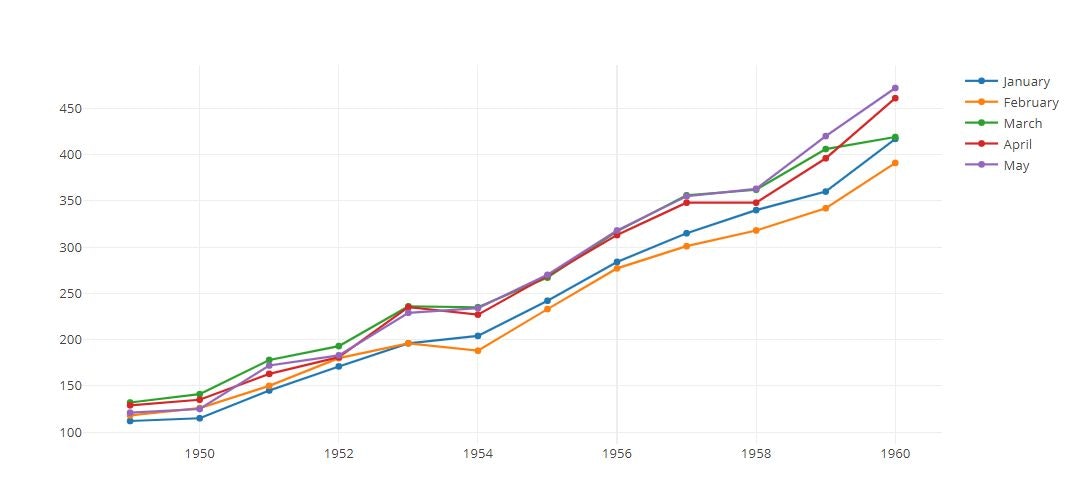
Line Plot
# Simple Line Plot
# Create a trace
trace1 = go.Scatter(
x = flights_data[flights_data['month'] == 'January']['year'],
y = flights_data[flights_data['month'] == 'January']['passengers'],
name='January'
)
trace2 = go.Scatter(
x = flights_data[flights_data['month'] == 'February']['year'],
y = flights_data[flights_data['month'] == 'February']['passengers'],
name='February'
)
trace3 = go.Scatter(
x = flights_data[flights_data['month'] == 'March']['year'],
y = flights_data[flights_data['month'] == 'March']['passengers'],
name='March'
)
trace4 = go.Scatter(
x = flights_data[flights_data['month'] == 'April']['year'],
y = flights_data[flights_data['month'] == 'April']['passengers'],
name='April'
)
trace5 = go.Scatter(
x = flights_data[flights_data['month'] == 'May']['year'],
y = flights_data[flights_data['month'] == 'May']['passengers'],
name='May'
)
tmp = [trace1,trace2,trace3,trace4,trace5]
py.offline.iplot(tmp)
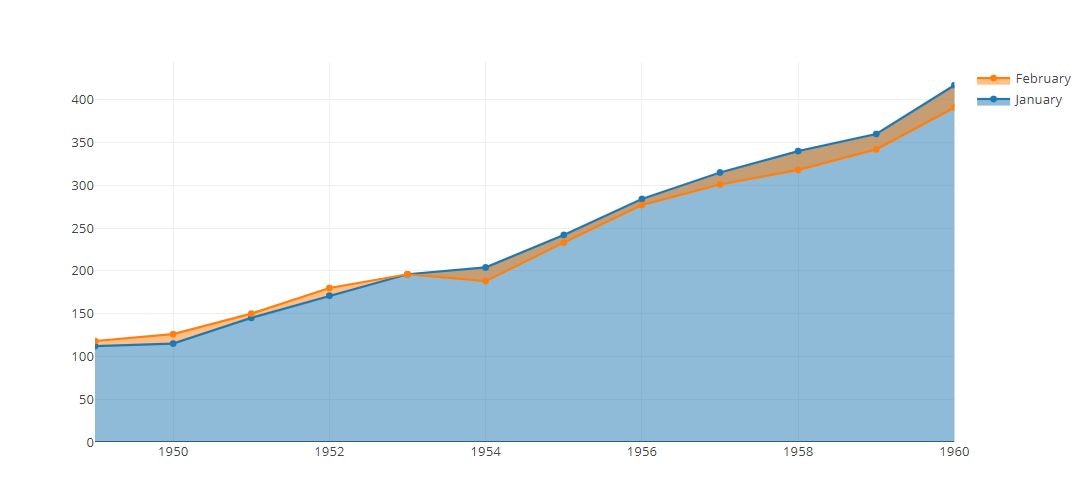
Filled Area Plot
trace1 = go.Scatter(
x = flights_data[flights_data['month'] == 'January']['year'],
y = flights_data[flights_data['month'] == 'January']['passengers'],
name='January',
fill='tozeroy'
)
trace2 = go.Scatter(
x = flights_data[flights_data['month'] == 'February']['year'],
y = flights_data[flights_data['month'] == 'February']['passengers'],
name='February',
fill='tonexty'
)
tmp = [trace1,trace2]
py.offline.iplot(tmp)
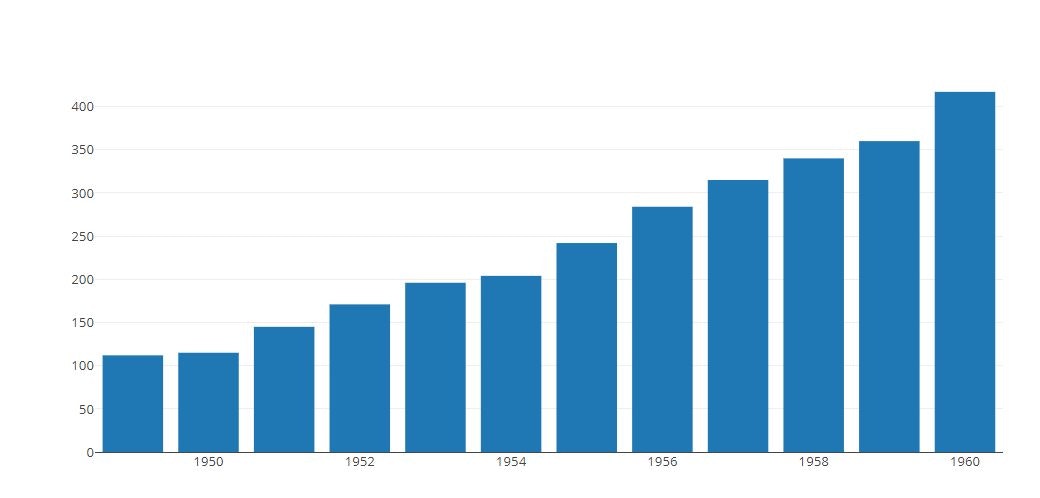
Bar Chart
# Basic
tmp = [go.Bar(
x=flights_data[flights_data['month'] == 'January']['year'],
y=flights_data[flights_data['month'] == 'January']['passengers'],
name='January'
)]
py.offline.iplot(tmp)
# Grouped Bar Chart
trace1 = go.Bar(
x=flights_data[flights_data['month'] == 'January']['year'],
y=flights_data[flights_data['month'] == 'January']['passengers'],
name='January'
)
trace2 = go.Bar(
x=flights_data[flights_data['month'] == 'February']['year'],
y=flights_data[flights_data['month'] == 'February']['passengers'],
name='February'
)
tmp = [trace1, trace2]
layout = go.Layout(
barmode='group'
)
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
# Stacked Bar Chart
trace1 = go.Bar(
x=flights_data[flights_data['month'] == 'January']['year'],
y=flights_data[flights_data['month'] == 'January']['passengers'],
name='January'
)
trace2 = go.Bar(
x=flights_data[flights_data['month'] == 'February']['year'],
y=flights_data[flights_data['month'] == 'February']['passengers'],
name='February'
)
tmp = [trace1, trace2]
layout = go.Layout(
barmode='stack'
)
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
# Horizontal Bar Chart
trace1 = go.Bar(
y=flights_data[flights_data['month'] == 'January']['year'],
x=flights_data[flights_data['month'] == 'January']['passengers'],
name='January',
orientation = 'h',
)
trace2 = go.Bar(
y=flights_data[flights_data['month'] == 'February']['year'],
x=flights_data[flights_data['month'] == 'February']['passengers'],
name='February',
orientation = 'h',
)
tmp = [trace1, trace2]
layout = go.Layout(
barmode='stack'
)
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
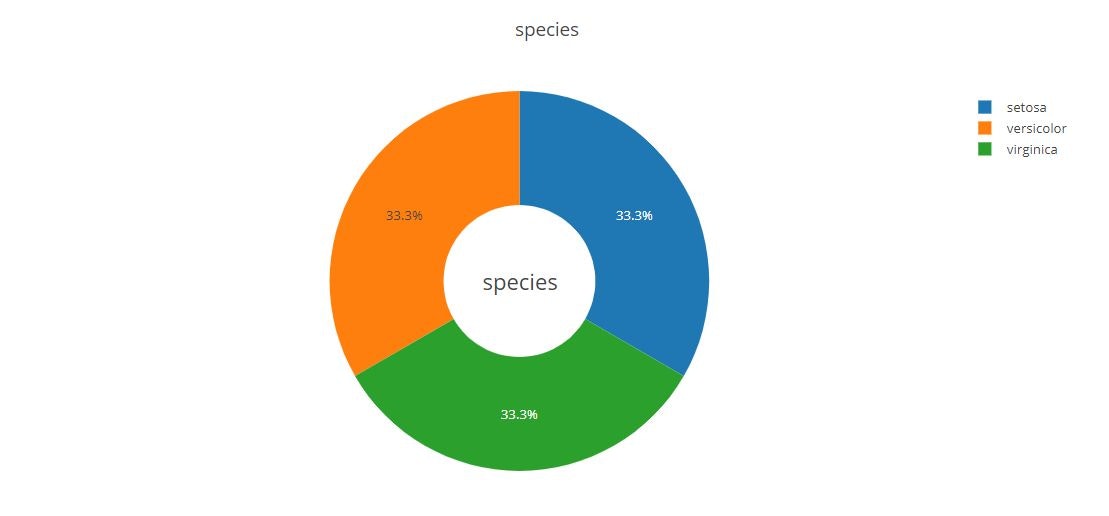
Pie Chart
# Basic
labels=['setosa','versicolor','virginica']
values=[len(iris_data[iris_data['species']=='setosa']),
len(iris_data[iris_data['species']=='versicolor']),
len(iris_data[iris_data['species']=='virginica'])]
trace=go.Pie(labels=labels, values=values)
py.offline.iplot([trace])
fig = {
"data": [
{
"values": [len(iris_data[iris_data['species']=='setosa']),
len(iris_data[iris_data['species']=='versicolor']),
len(iris_data[iris_data['species']=='virginica'])],
"labels": ['setosa','versicolor','virginica'],
"domain": {"column": 0},
"name": "species",
"hoverinfo":"label+percent+name",
"hole": .4,
"type": "pie"
}],
"layout": {
"title":"species",
"grid": {"rows": 1, "columns": 1},
"annotations": [
{
"font": {
"size": 20
},
"showarrow": False,
"text": "species",
"x": 0.5,
"y": 0.5
}
]
}
}
py.offline.iplot(fig)
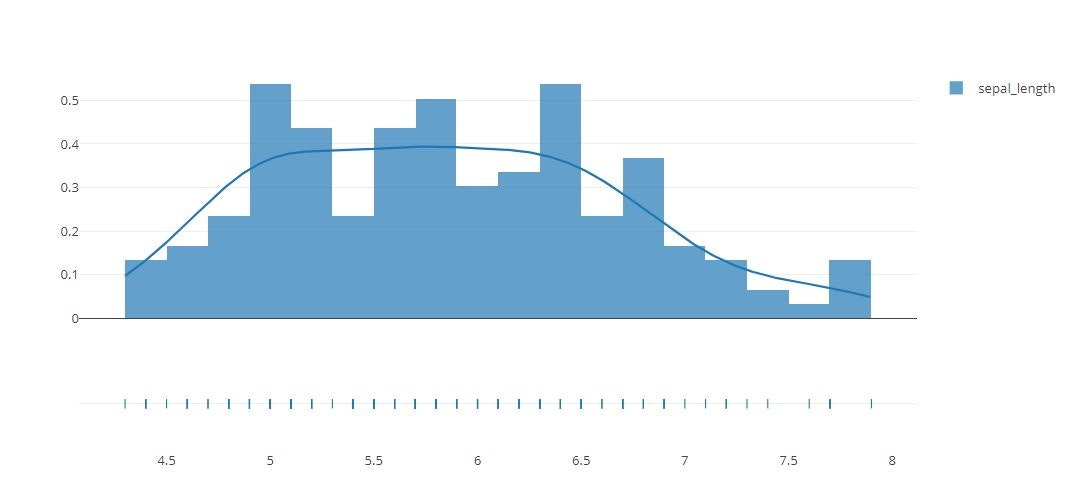
Distplot
x = iris_data['sepal_length']
hist_data = [x]
group_labels = ['sepal_length']
fig = ff.create_distplot(hist_data, group_labels,bin_size=.2)
py.offline.iplot(fig)
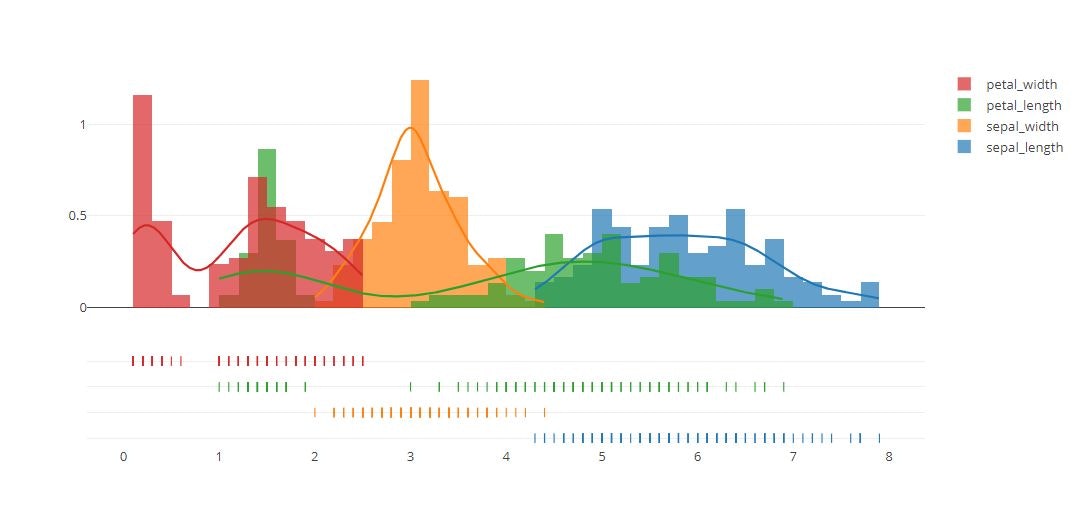
# Add histogram data
x1 = iris_data['sepal_length']
x2 = iris_data['sepal_width']
x3 = iris_data['petal_length']
x4 = iris_data['petal_width']
# Group data together
hist_data = [x1, x2, x3, x4]
group_labels = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
# Create distplot with custom bin_size
fig = ff.create_distplot(hist_data, group_labels, bin_size=.2)
# Plot!
py.offline.iplot(fig)
# Create distplot with custom bin_size
fig = ff.create_distplot(hist_data, group_labels, bin_size=[.1, .25, .5, 1])
# Plot!
py.offline.iplot(fig)
rug_text_one = ['a', 'b', 'c', 'd', 'e',
'f', 'g', 'h', 'i', 'j',
'k', 'l', 'm', 'n', 'o',
'p', 'q', 'r', 's', 't',
'u', 'v', 'w', 'x', 'y', 'z']
rug_text_two = ['aa', 'bb', 'cc', 'dd', 'ee',
'ff', 'gg', 'hh', 'ii', 'jj',
'kk', 'll', 'mm', 'nn', 'oo',
'pp', 'qq', 'rr', 'ss', 'tt',
'uu', 'vv', 'ww', 'xx', 'yy', 'zz']
rug_text_three = ['aaa', 'bbb', 'ccc', 'ddd', 'eee',
'fff', 'ggg', 'hhh', 'iii', 'jjj',
'kkk', 'lll', 'mmm', 'nnn', 'ooo',
'ppp', 'qqq', 'rrr', 'sss', 'ttt',
'uuu', 'vvv', 'www', 'xxx', 'yyy', 'zzz']
rug_text_four = ['aaaa', 'bbbb', 'cccc', 'dddd', 'eeee',
'ffff', 'gggg', 'hhhh', 'iiii', 'jjjj',
'kkkk', 'llll', 'mmmm', 'nnnn', 'oooo',
'pppp', 'qqqq', 'rrrr', 'ssss', 'tttt',
'uuuu', 'vvvv', 'wwww', 'xxxx', 'yyyy', 'zzzz']
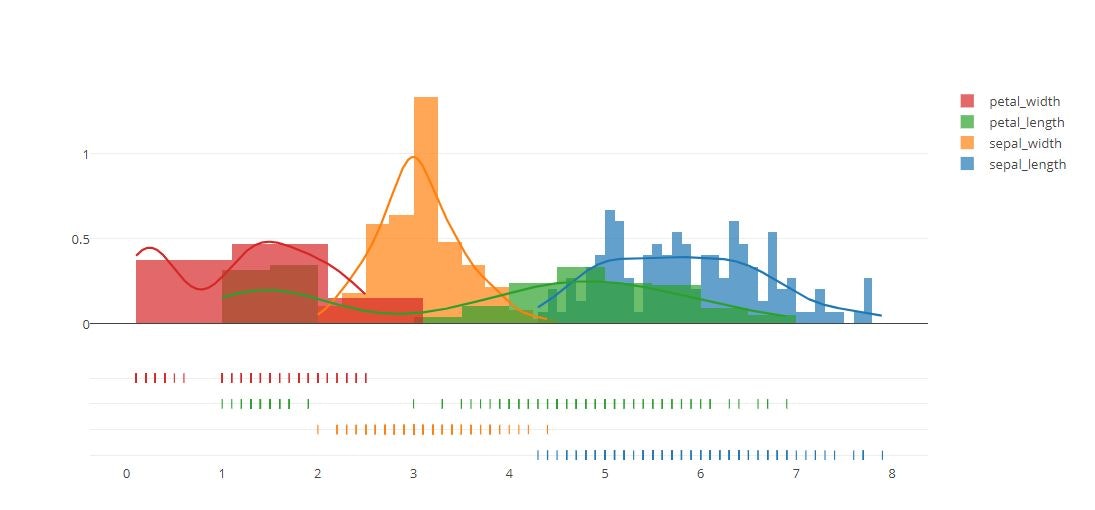
rug_text = [rug_text_one, rug_text_two,rug_text_three,rug_text_four]
colors = ['rgb(0, 0, 100)', 'rgb(0, 100, 100)','rgb(100, 100, 100)','rgb(100, 100, 200)']
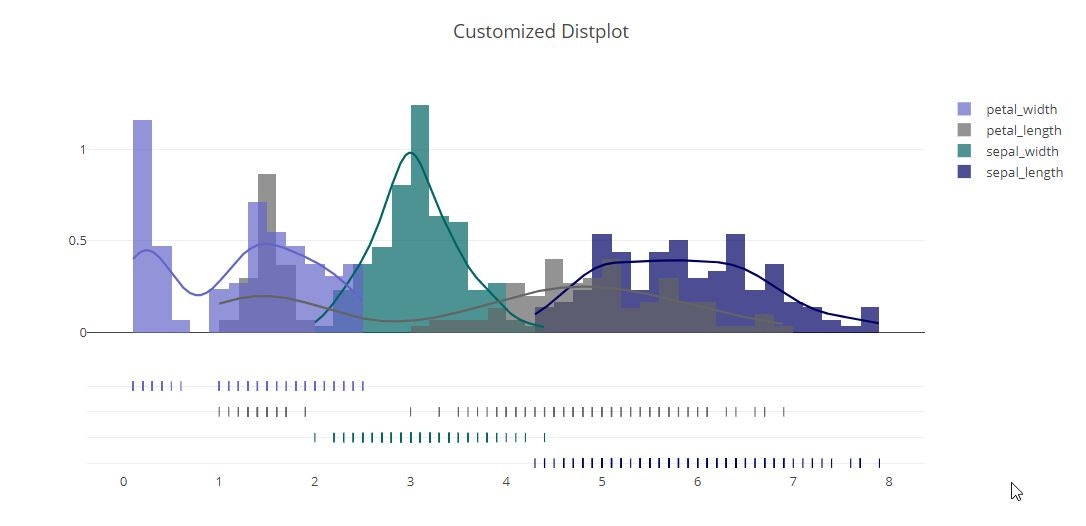
fig = ff.create_distplot(
hist_data, group_labels, bin_size=.2,
rug_text=rug_text, colors=colors)
fig['layout'].update(title='Customized Distplot')
# Plot!
py.offline.iplot(fig)
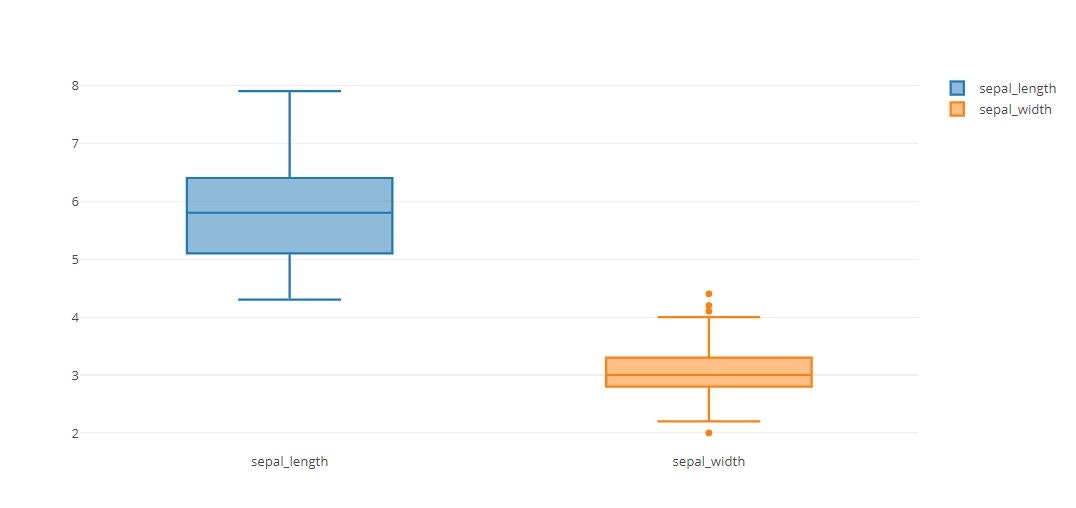
trace0 = go.Box(
y=iris_data['sepal_length'],
name='sepal_length'
)
trace1 = go.Box(
y=iris_data['sepal_width'],
name='sepal_width'
)
tmp = [trace0, trace1]
py.offline.iplot(tmp)
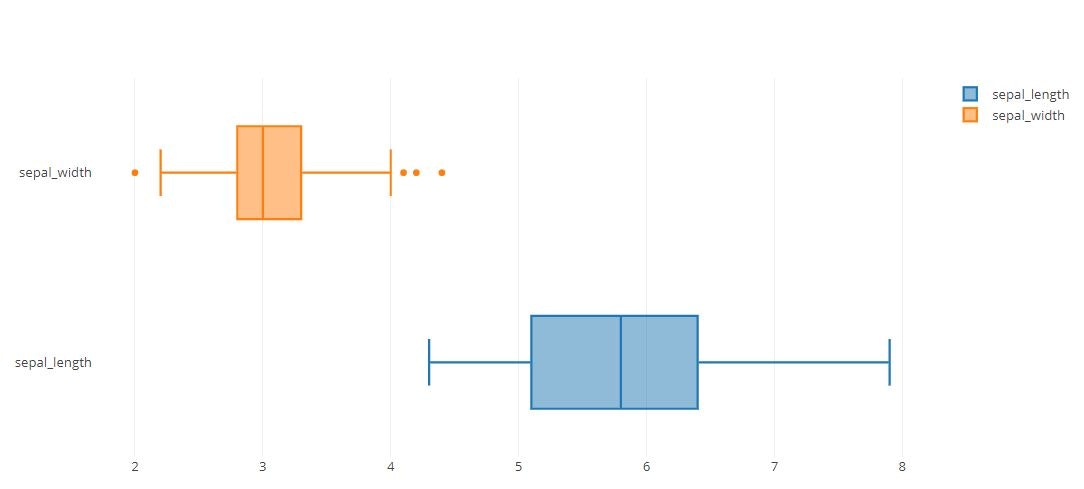
Boxplot
trace0 = go.Box(
x=iris_data['sepal_length'],
name='sepal_length'
)
trace1 = go.Box(
x=iris_data['sepal_width'],
name='sepal_width'
)
tmp = [trace0, trace1]
py.offline.iplot(tmp)
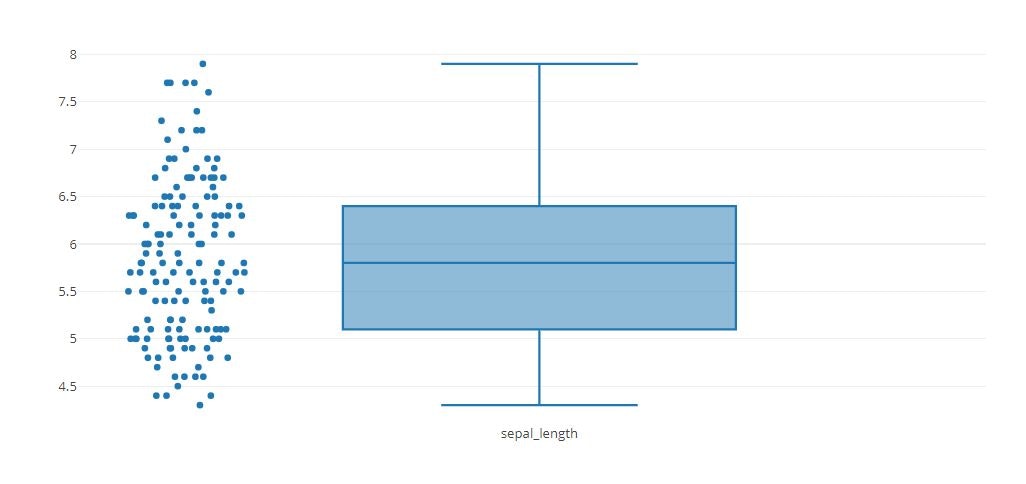
trace0 = go.Box(
y=iris_data['sepal_length'],
name='sepal_length',
boxpoints='all',
jitter=0.3,
pointpos=-1.8
)
tmp = [trace0]
py.offline.iplot(tmp)
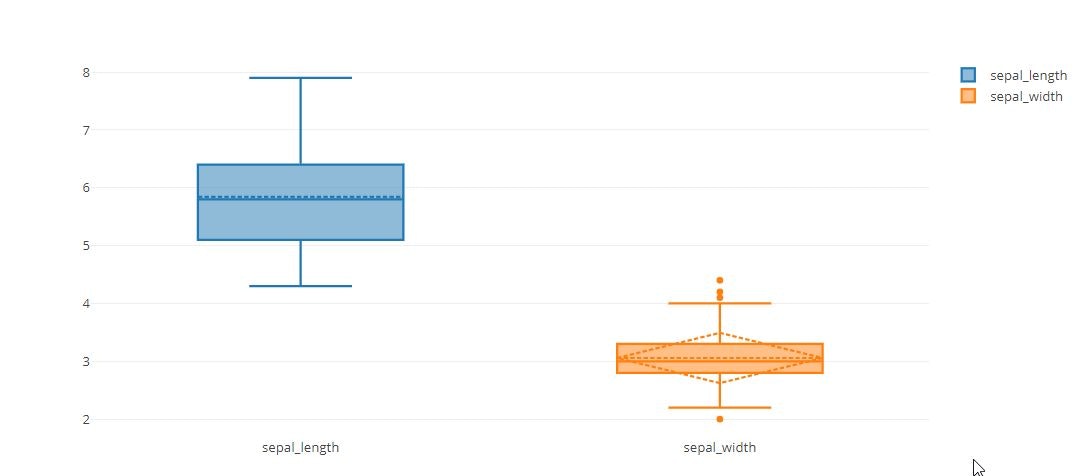
trace0 = go.Box(
y=iris_data['sepal_length'],
name='sepal_length',
marker = dict(
color = 'rgb(214, 12, 140)',
)
)
trace1 = go.Box(
y=iris_data['sepal_width'],
name='sepal_width',
marker = dict(
color = 'rgb(0, 128, 128)',
)
)
tmp = [trace0, trace1]
py.offline.iplot(tmp)
trace0 = go.Box(
y=iris_data['sepal_length'],
name='sepal_length',
boxmean=True
)
trace1 = go.Box(
y=iris_data['sepal_width'],
name='sepal_width',
boxmean='sd'
)
tmp = [trace0, trace1]
py.offline.iplot(tmp)
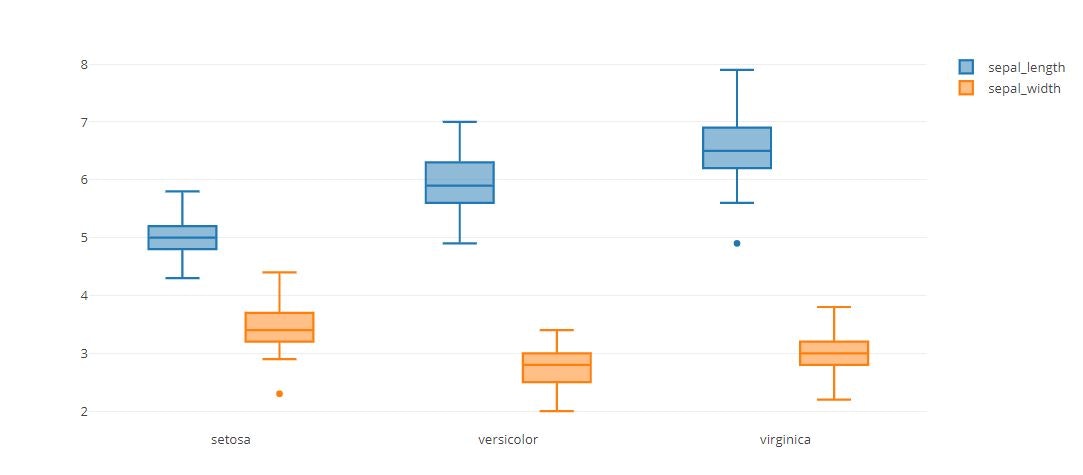
trace0 = go.Box(
y=iris_data['sepal_length'],
name='sepal_length',
x=iris_data['species']
)
trace1 = go.Box(
y=iris_data['sepal_width'],
name='sepal_width',
x=iris_data['species']
)
tmp = [trace0, trace1]
layout = go.Layout(
boxmode='group'
)
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
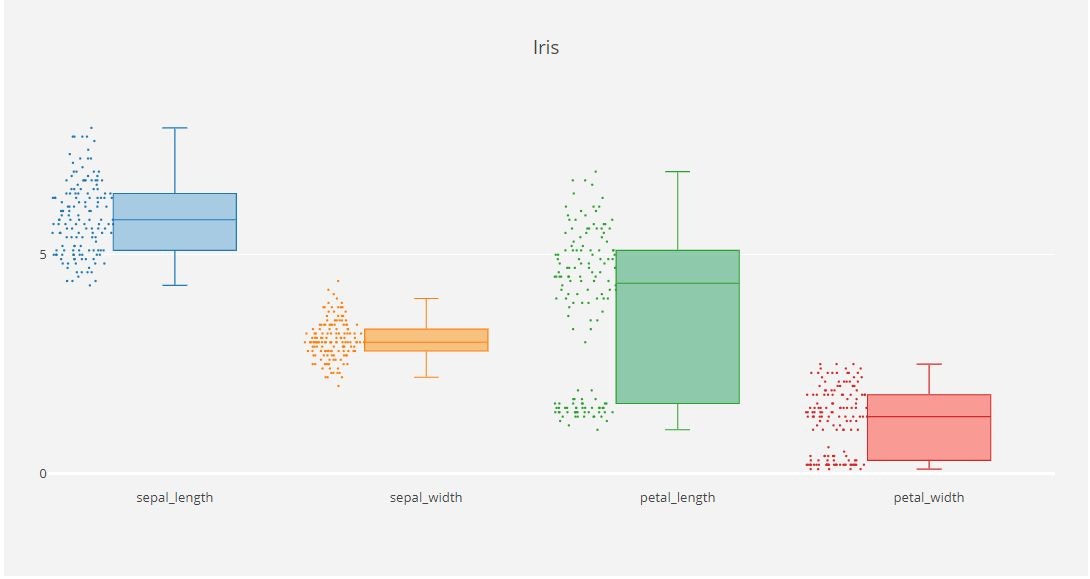
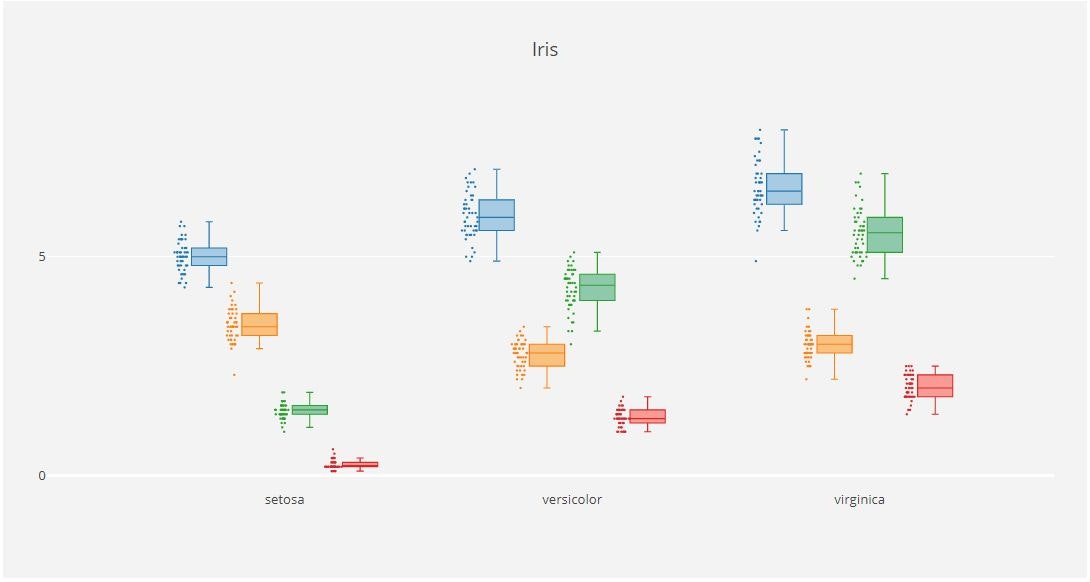
x_data = ['sepal_length', 'sepal_width',
'petal_length', 'petal_width']
y0 = iris_data['sepal_length']
y1 = iris_data['sepal_width']
y2 = iris_data['petal_length']
y3 = iris_data['petal_width']
y_data = [y0,y1,y2,y3]
colors = ['rgba(93, 164, 214, 0.5)', 'rgba(255, 144, 14, 0.5)', 'rgba(44, 160, 101, 0.5)', 'rgba(255, 65, 54, 0.5)']
traces = []
for xd, yd, cls in zip(x_data, y_data, colors):
traces.append(go.Box(
y=yd,
name=xd,
boxpoints='all',
jitter=0.5,
whiskerwidth=0.2,
fillcolor=cls,
marker=dict(
size=2,
),
line=dict(width=1),
))
layout = go.Layout(
title='Iris',
yaxis=dict(
autorange=True,
showgrid=True,
zeroline=True,
dtick=5,
gridcolor='rgb(255, 255, 255)',
gridwidth=1,
zerolinecolor='rgb(255, 255, 255)',
zerolinewidth=2,
),
margin=dict(
l=40,
r=30,
b=80,
t=100,
),
paper_bgcolor='rgb(243, 243, 243)',
plot_bgcolor='rgb(243, 243, 243)',
showlegend=False
)
fig = go.Figure(data=traces, layout=layout)
py.offline.iplot(fig)
x_data = ['sepal_length', 'sepal_width',
'petal_length', 'petal_width']
y0 = iris_data['sepal_length']
y1 = iris_data['sepal_width']
y2 = iris_data['petal_length']
y3 = iris_data['petal_width']
y_data = [y0,y1,y2,y3]
colors = ['rgba(93, 164, 214, 0.5)', 'rgba(255, 144, 14, 0.5)', 'rgba(44, 160, 101, 0.5)', 'rgba(255, 65, 54, 0.5)']
traces = []
for xd, yd, cls in zip(x_data, y_data, colors):
traces.append(go.Box(
y=yd,
x=iris_data['species'],
name=xd,
boxpoints='all',
jitter=0.5,
whiskerwidth=0.2,
fillcolor=cls,
marker=dict(
size=2,
),
line=dict(width=1),
))
layout = go.Layout(
title='Iris',
yaxis=dict(
autorange=True,
showgrid=True,
zeroline=True,
dtick=5,
gridcolor='rgb(255, 255, 255)',
gridwidth=1,
zerolinecolor='rgb(255, 255, 255)',
zerolinewidth=2,
),
margin=dict(
l=40,
r=30,
b=80,
t=100,
),
paper_bgcolor='rgb(243, 243, 243)',
plot_bgcolor='rgb(243, 243, 243)',
showlegend=False,
boxmode='group'
)
fig = go.Figure(data=traces, layout=layout)
py.offline.iplot(fig)
Histgram
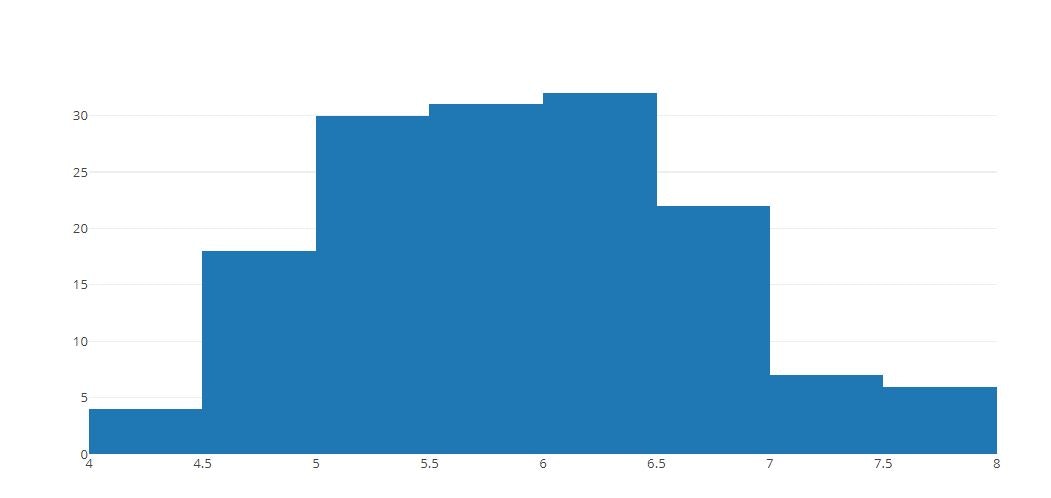
# Basic Histogram
x = iris_data['sepal_length']
tmp = [go.Histogram(x=x)]
py.offline.iplot(tmp)
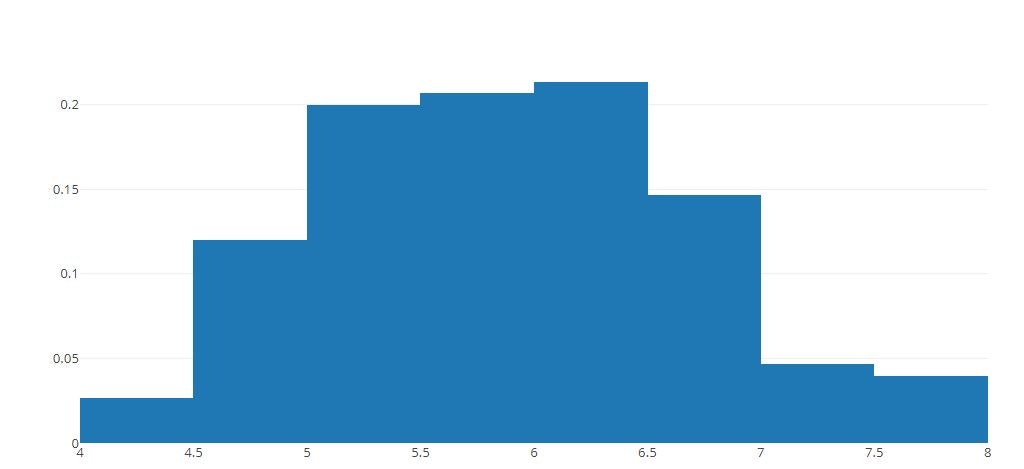
# Normalized Histogram
tmp = [go.Histogram(x=x,histnorm='probability')]
py.offline.iplot(tmp)
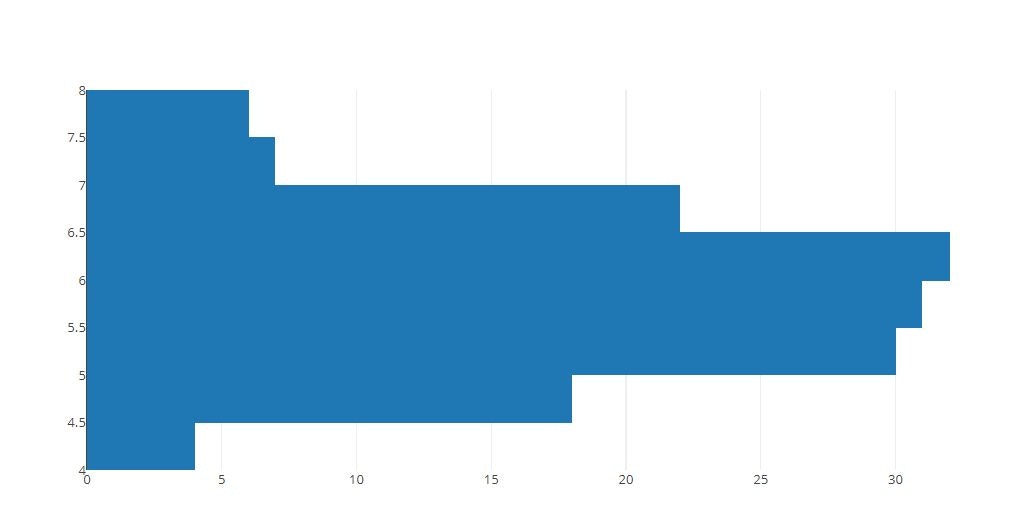
# Horizontal Histogram
tmp = [go.Histogram(y=x)]
py.offline.iplot(tmp)
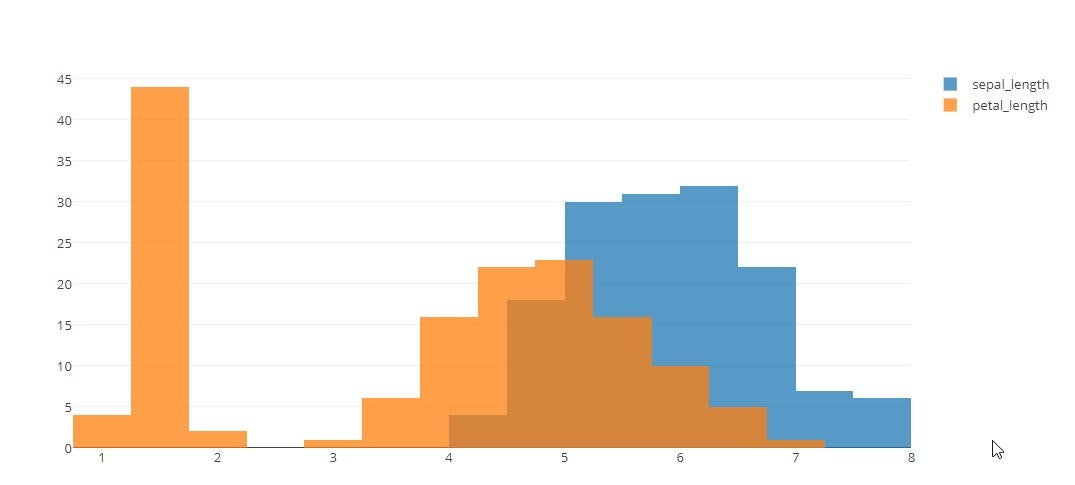
# Overlaid Histogram
x0 = iris_data['sepal_length']
x1 = iris_data['petal_length']
trace1 = go.Histogram(
x=x0,
opacity=0.75,
name='sepal_length'
)
trace2 = go.Histogram(
x=x1,
opacity=0.75,
name='petal_length'
)
tmp = [trace1, trace2]
layout = go.Layout(barmode='overlay')
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
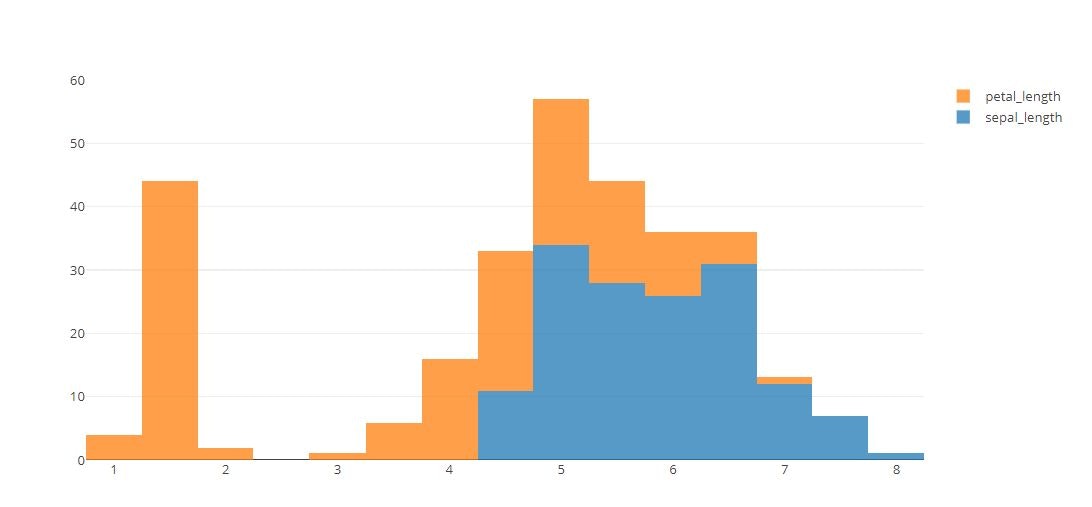
# Stacked Histograms
layout = go.Layout(barmode='stack')
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
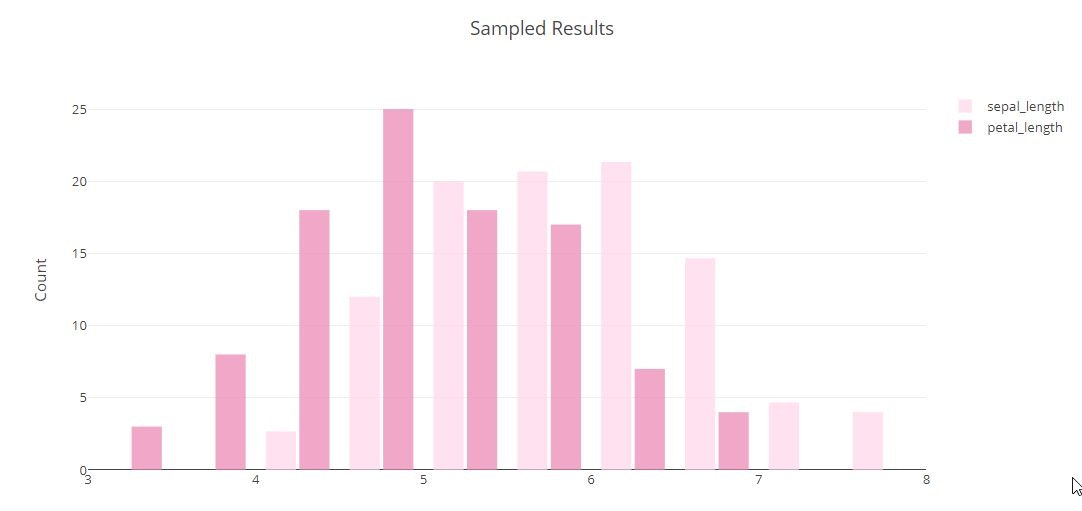
# Styled Histogram
x0 = iris_data['sepal_length']
x1 = iris_data['petal_length']
trace1 = go.Histogram(
x=x0,
histnorm='percent',
name='sepal_length',
xbins=dict(
start=3.0,
end=8.0,
size=0.5
),
marker=dict(
color='#FFD7E9',
),
opacity=0.75
)
trace2 = go.Histogram(
x=x1,
name='petal_length',
xbins=dict(
start=3.0,
end=8.0,
size=0.5
),
marker=dict(
color='#EB89B5'
),
opacity=0.75
)
tmp = [trace1, trace2]
layout = go.Layout(
title='Sampled Results',
xaxis=dict(
title='Value'
),
yaxis=dict(
title='Count'
),
bargap=0.2,
bargroupgap=0.1
)
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
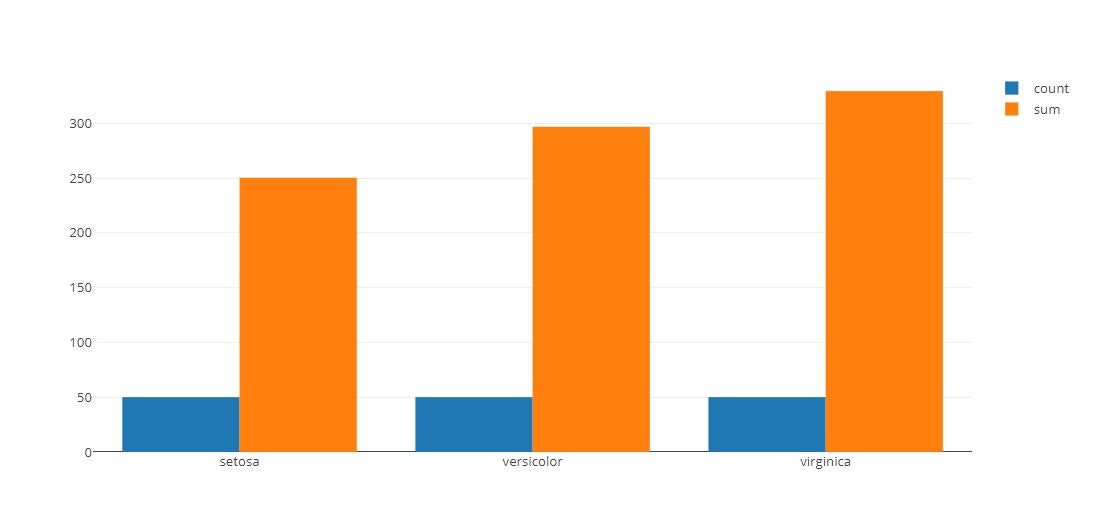
# Specify Binning Function
x = iris_data['species']
y = iris_data['sepal_length']
tmp = [
go.Histogram(
histfunc = "count",
y = y,
x = x,
name = "count"
),
go.Histogram(
histfunc = "sum",
y = y,
x = x,
name = "sum"
)
]
py.offline.iplot(tmp)
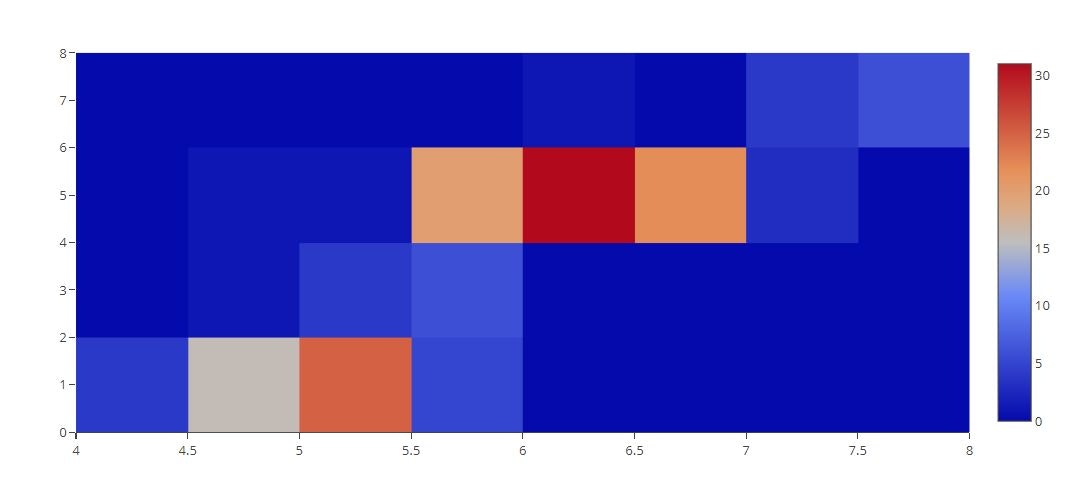
2D Histogram
# 2D Histogram of a Bivariate Normal Distribution
x = iris_data['sepal_length']
y = iris_data['petal_length']
tmp = [
go.Histogram2d(
x=x,
y=y
)
]
py.offline.iplot(tmp)
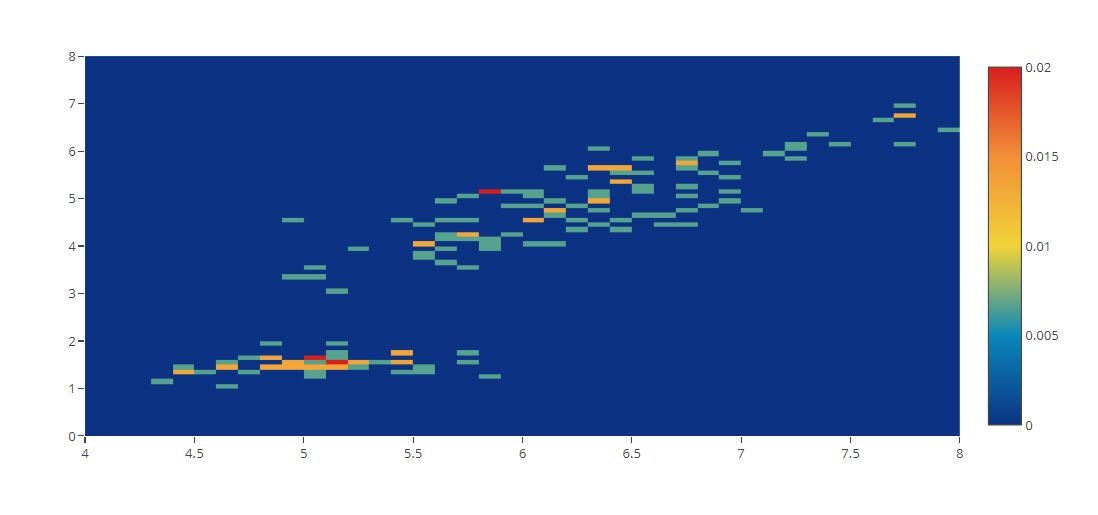
# 2D Histogram Binning and Styling Options
x = iris_data['sepal_length']
y = iris_data['petal_length']
tmp = [
go.Histogram2d(x=x, y=y, histnorm='probability',
autobinx=False,
xbins=dict(start=4, end=8, size=0.1),
autobiny=False,
ybins=dict(start=0, end=8, size=0.1),
colorscale=[[0, 'rgb(12,51,131)'], [0.25, 'rgb(10,136,186)'], [0.5, 'rgb(242,211,56)'], [0.75, 'rgb(242,143,56)'], [1, 'rgb(217,30,30)']]
)
]
py.offline.iplot(tmp)
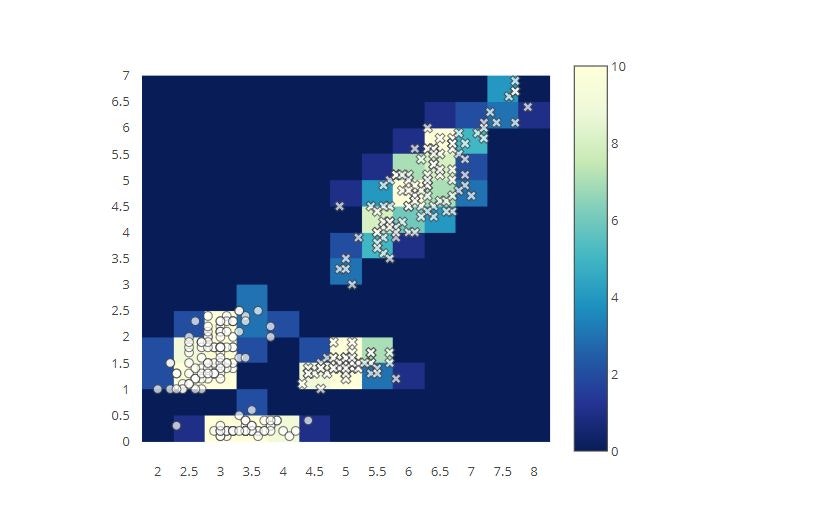
# 2D Histogram Overlaid with a Scatter Chart
x0 = iris_data['sepal_length']
y0 = iris_data['petal_length']
x1 = iris_data['sepal_width']
y1 = iris_data['petal_width']
x = np.concatenate([x0, x1])
y = np.concatenate([y0, y1])
trace1 = go.Scatter(
x=x0,
y=y0,
mode='markers',
showlegend=False,
marker=dict(
symbol='x',
opacity=0.7,
color='white',
size=8,
line=dict(width=1),
)
)
trace2 = go.Scatter(
x=x1,
y=y1,
mode='markers',
showlegend=False,
marker=dict(
symbol='circle',
opacity=0.7,
color='white',
size=8,
line=dict(width=1),
)
)
trace3 = go.Histogram2d(
x=x,
y=y,
colorscale='YlGnBu',
zmax=10,
nbinsx=14,
nbinsy=14,
zauto=False,
)
layout = go.Layout(
xaxis=dict( ticks='', showgrid=False, zeroline=False, nticks=20 ),
yaxis=dict( ticks='', showgrid=False, zeroline=False, nticks=20 ),
autosize=False,
height=550,
width=550,
hovermode='closest',
)
tmp = [trace1, trace2, trace3]
fig = go.Figure(data=tmp, layout=layout)
py.offline.iplot(fig)
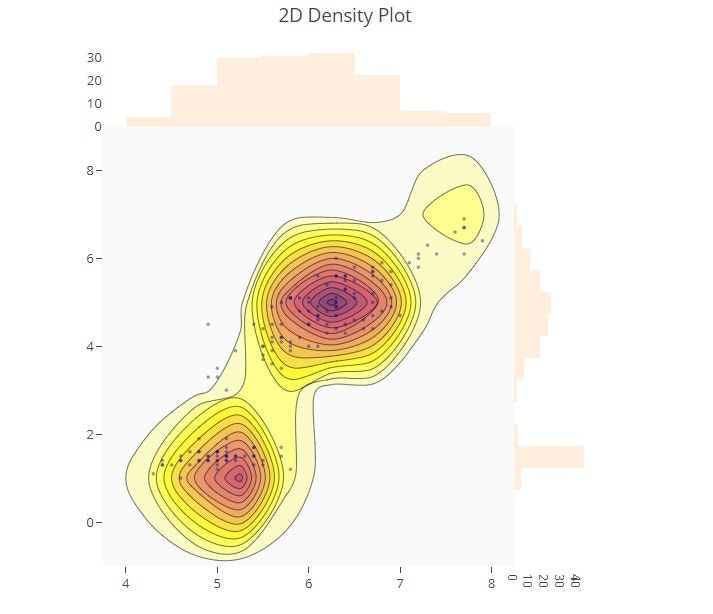
2D Density Plot
# 2D Histogram Contour Plot with Histogram Subplots
t = np.linspace(-1, 1.2, 2000)
x = iris_data['sepal_length']
y = iris_data['petal_length']
colorscale = ['#7A4579', '#D56073', 'rgb(236,158,105)', (1, 1, 0.2), (0.98,0.98,0.98)]
fig = ff.create_2d_density(
x, y, colorscale=colorscale,
hist_color='rgb(255, 237, 222)', point_size=3
)
py.offline.iplot(fig)
Facet and Trellis Plot
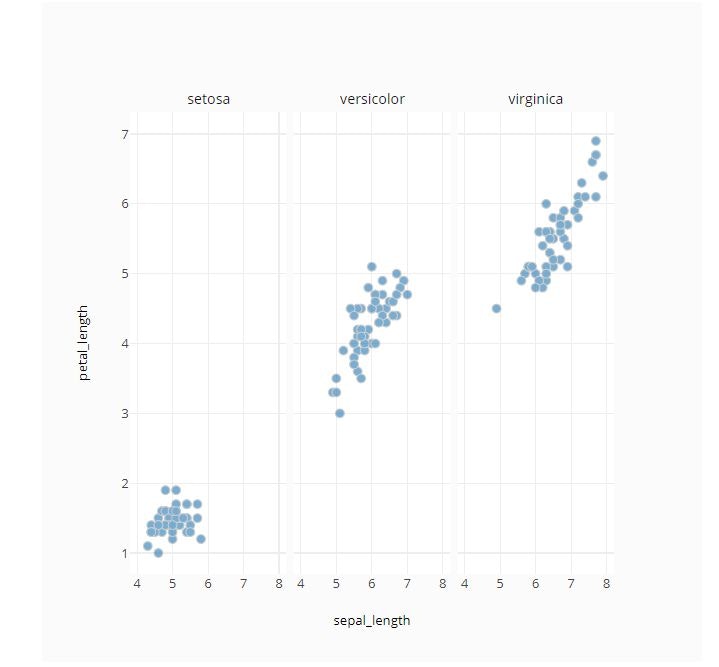
# Facet by Column
fig = ff.create_facet_grid(
iris_data,
x='sepal_length',
y='petal_length',
facet_col='species',
)
py.offline.iplot(fig)
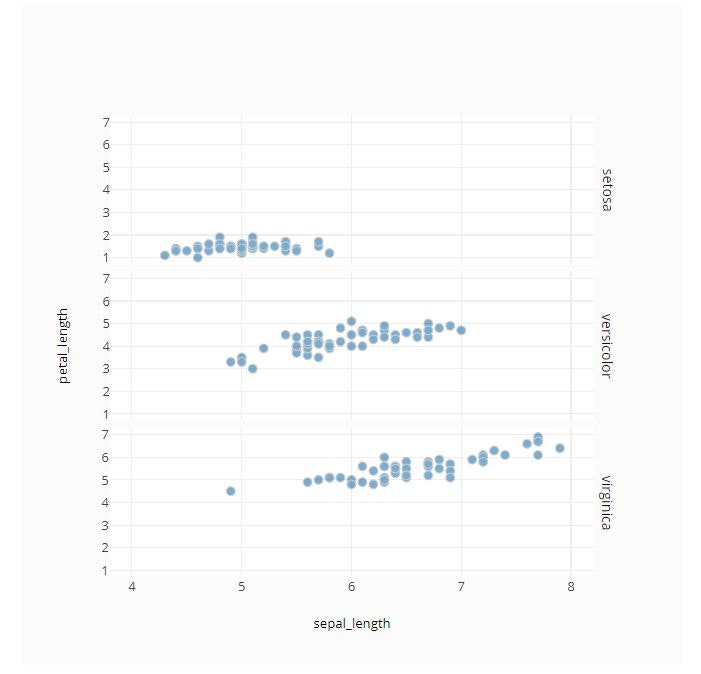
# Facet by Row
fig = ff.create_facet_grid(
iris_data,
x='sepal_length',
y='petal_length',
facet_row='species',
)
py.offline.iplot(fig)
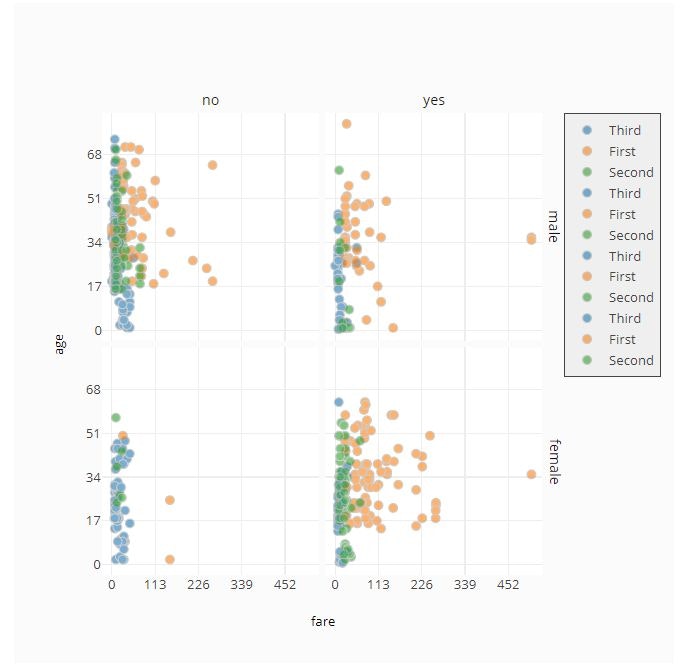
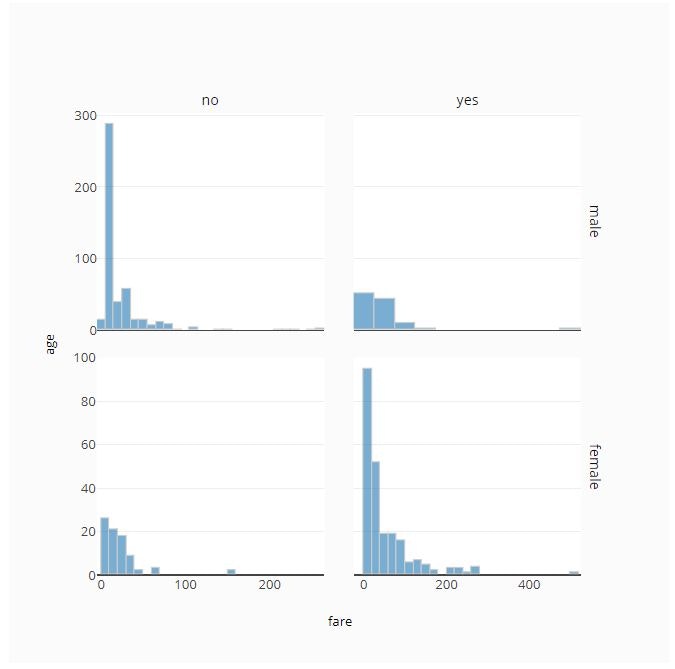
# Facet by Row and Column
fig = ff.create_facet_grid(
titanic_data,
x='fare',
y='age',
facet_row='sex',
facet_col='alive',
color_name='class',
color_is_cat=True,
)
py.offline.iplot(fig)
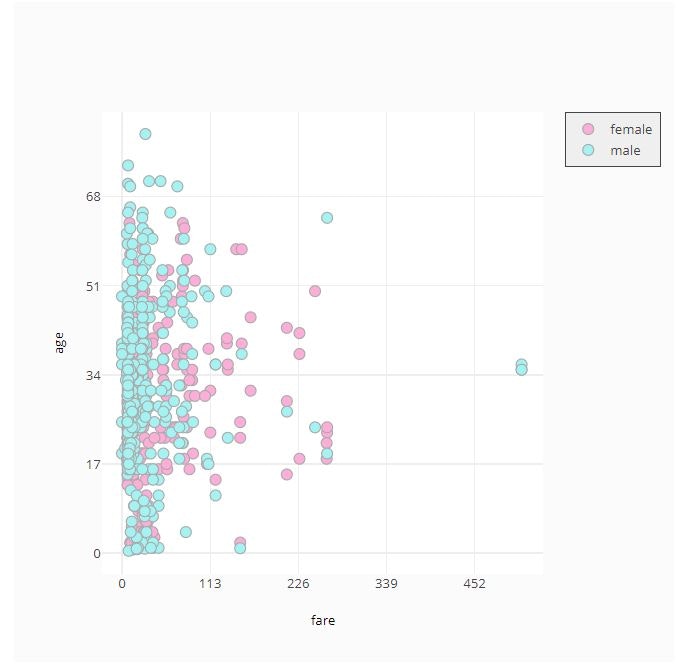
# Custom Colormap
fig = ff.create_facet_grid(
titanic_data,
x='fare',
y='age',
color_name='sex',
show_boxes=False,
marker={'size': 10, 'opacity': 1.0},
colormap={'male': 'rgb(165, 242, 242)', 'female': 'rgb(253, 174, 216)'}
)
py.offline.iplot(fig)
# Plot with Histogram Traces
fig = ff.create_facet_grid(
titanic_data,
x='fare',
y='age',
facet_row='sex',
facet_col='alive',
trace_type='histogram',
)
py.offline.iplot(fig)
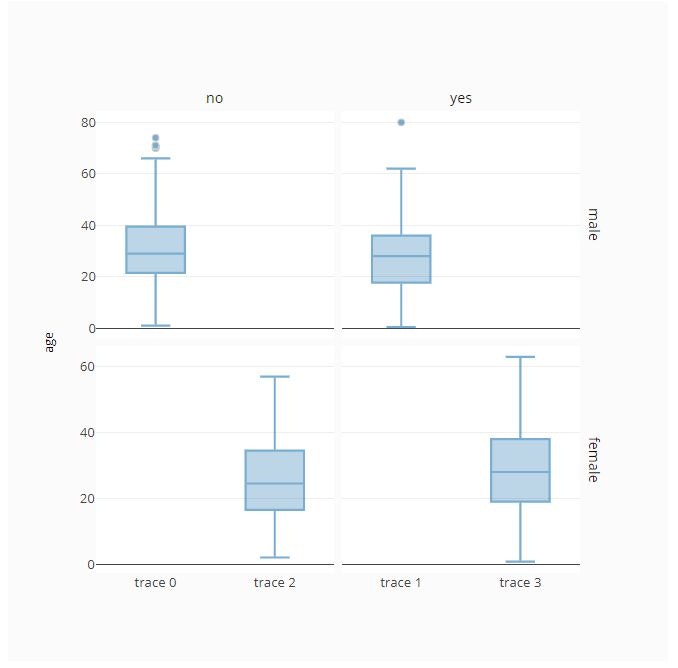
# Plot with BoxPlot Traces
fig = ff.create_facet_grid(
titanic_data,
y='age',
facet_row='sex',
facet_col='alive',
trace_type='box',
)
py.offline.iplot(fig)
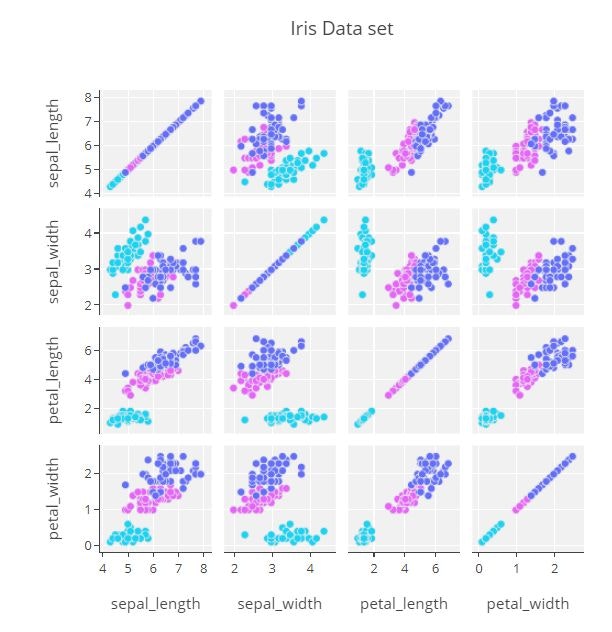
Scatterplot Matrix
classes=np.unique(iris_data['species'].values).tolist()
class_code={classes[k]: k for k in range(3)}
color_vals=[class_code[cl] for cl in iris_data['species']]
text=[iris_data.loc[ k, 'species'] for k in range(len(iris_data))]
pl_colorscale=[[0.0, '#19d3f3'],
[0.333, '#19d3f3'],
[0.333, '#e763fa'],
[0.666, '#e763fa'],
[0.666, '#636efa'],
[1, '#636efa']]
trace1 = go.Splom(dimensions=[dict(label='sepal_length',
values=iris_data['sepal_length']),
dict(label='sepal_width',
values=iris_data['sepal_width']),
dict(label='petal_length',
values=iris_data['petal_length']),
dict(label='petal_width',
values=iris_data['petal_width'])],
text=text,
marker=dict(color=color_vals,
size=7,
colorscale=pl_colorscale,
showscale=False,
line=dict(width=0.5,
color='rgb(230,230,230)'))
)
axis = dict(showline=True,
zeroline=False,
gridcolor='#fff',
ticklen=4)
layout = go.Layout(
title='Iris Data set',
dragmode='select',
width=600,
height=600,
autosize=False,
hovermode='closest',
plot_bgcolor='rgba(240,240,240, 0.95)',
xaxis1=dict(axis),
xaxis2=dict(axis),
xaxis3=dict(axis),
xaxis4=dict(axis),
yaxis1=dict(axis),
yaxis2=dict(axis),
yaxis3=dict(axis),
yaxis4=dict(axis)
)
fig1 = dict(data=[trace1], layout=layout)
py.offline.iplot(fig1)
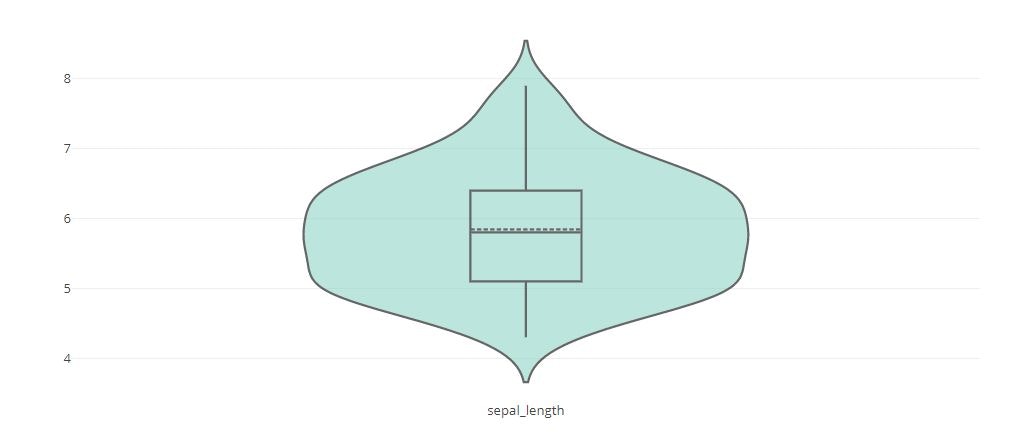
Violin Plot
# Basic Violin Plot
fig = {
"data": [{
"type": 'violin',
"y": iris_data['sepal_length'],
"box": {
"visible": True
},
"line": {
"color": 'black'
},
"meanline": {
"visible": True
},
"fillcolor": '#8dd3c7',
"opacity": 0.6,
"x0": 'sepal_length'
}],
"layout" : {
"title": "",
"yaxis": {
"zeroline": False,
}
}
}
py.offline.iplot(fig)
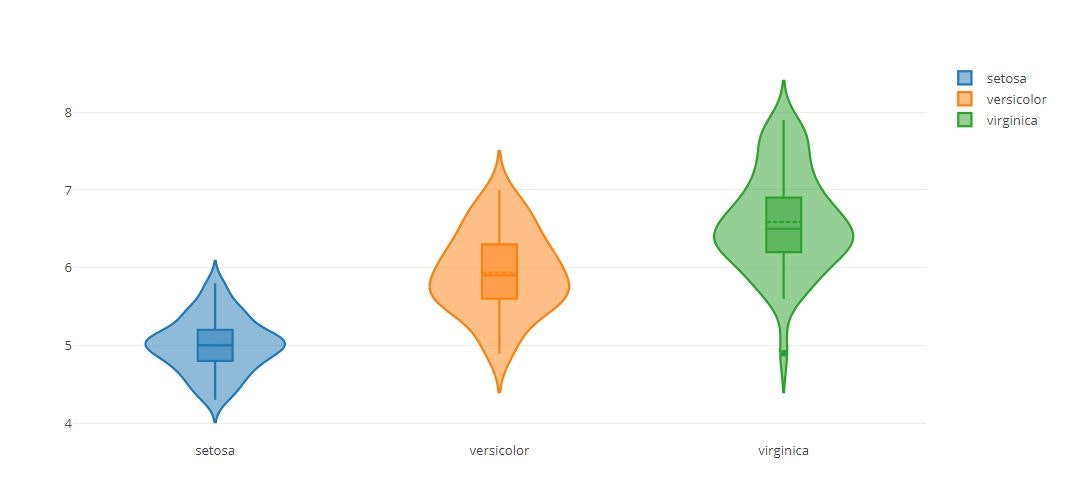
# Multiple Traces
tmp = []
for i in range(0,len(pd.unique(iris_data['species']))):
trace = {
"type": 'violin',
"x": iris_data['species'][iris_data['species'] == pd.unique(iris_data['species'])[i]],
"y": iris_data['sepal_length'][iris_data['species'] == pd.unique(iris_data['species'])[i]],
"name": pd.unique(iris_data['species'])[i],
"box": {
"visible": True
},
"meanline": {
"visible": True
}
}
tmp.append(trace)
fig = {
"data": tmp,
"layout" : {
"title": "",
"yaxis": {
"zeroline": False,
}
}
}
py.offline.iplot(fig)
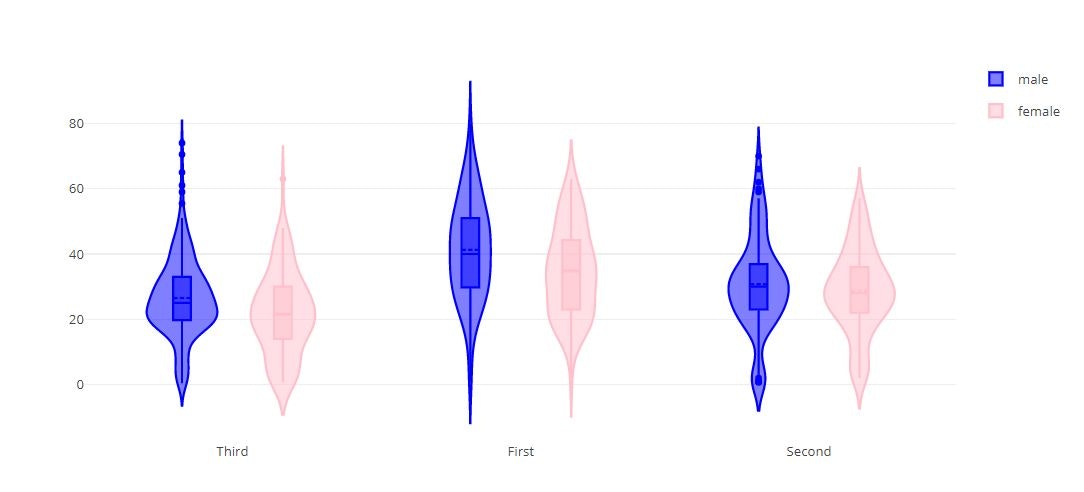
# Grouped Violin Plot
fig = {
"data": [
{
"type": 'violin',
"x": titanic_data['class'][ titanic_data['sex'] == 'male' ],
"y": titanic_data['age'] [ titanic_data['sex'] == 'male' ],
"legendgroup": 'male',
"scalegroup": 'male',
"name": 'male',
"box": {
"visible": True
},
"meanline": {
"visible": True
},
"line": {
"color": 'blue'
}
},
{
"type": 'violin',
"x": titanic_data['class'][ titanic_data['sex'] == 'female' ],
"y": titanic_data['age'] [ titanic_data['sex'] == 'female' ],
"legendgroup": 'female',
"scalegroup": 'female',
"name": 'female',
"box": {
"visible": True
},
"meanline": {
"visible": True
},
"line": {
"color": 'pink'
}
}
],
"layout" : {
"yaxis": {
"zeroline": False,
},
"violinmode": "group"
}
}
py.offline.iplot(fig)
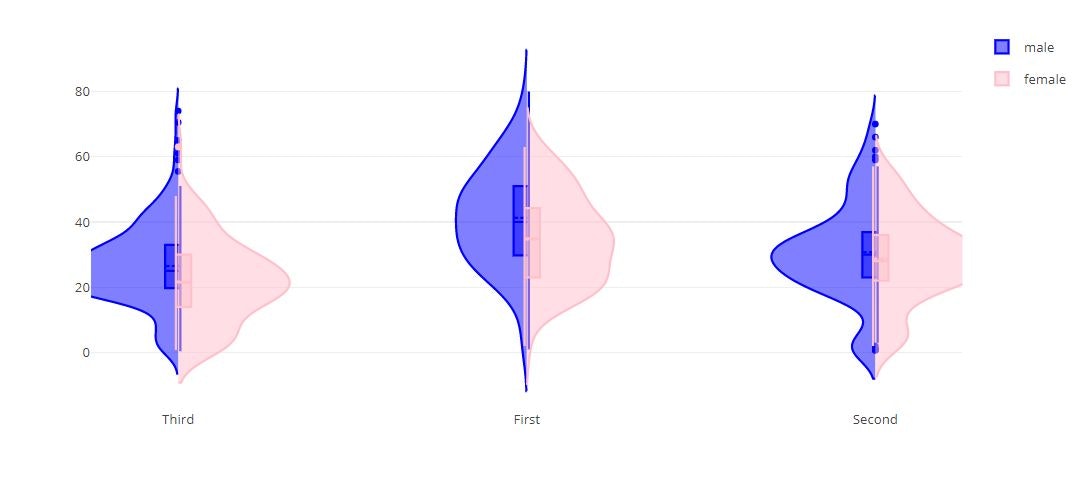
# Split Violin Plot
fig = {
"data": [
{
"type": 'violin',
"x": titanic_data['class'][ titanic_data['sex'] == 'male' ],
"y": titanic_data['age'] [ titanic_data['sex'] == 'male' ],
"legendgroup": 'male',
"scalegroup": 'male',
"name": 'male',
"side": 'negative',
"box": {
"visible": True
},
"meanline": {
"visible": True
},
"line": {
"color": 'blue'
}
},
{
"type": 'violin',
"x": titanic_data['class'][ titanic_data['sex'] == 'female' ],
"y": titanic_data['age'] [ titanic_data['sex'] == 'female' ],
"legendgroup": 'female',
"scalegroup": 'female',
"name": 'female',
"side": 'positive',
"box": {
"visible": True
},
"meanline": {
"visible": True
},
"line": {
"color": 'pink'
}
}
],
"layout" : {
"yaxis": {
"zeroline": False,
},
"violingap": 0,
"violinmode": "overlay"
}
}
py.offline.iplot(fig)
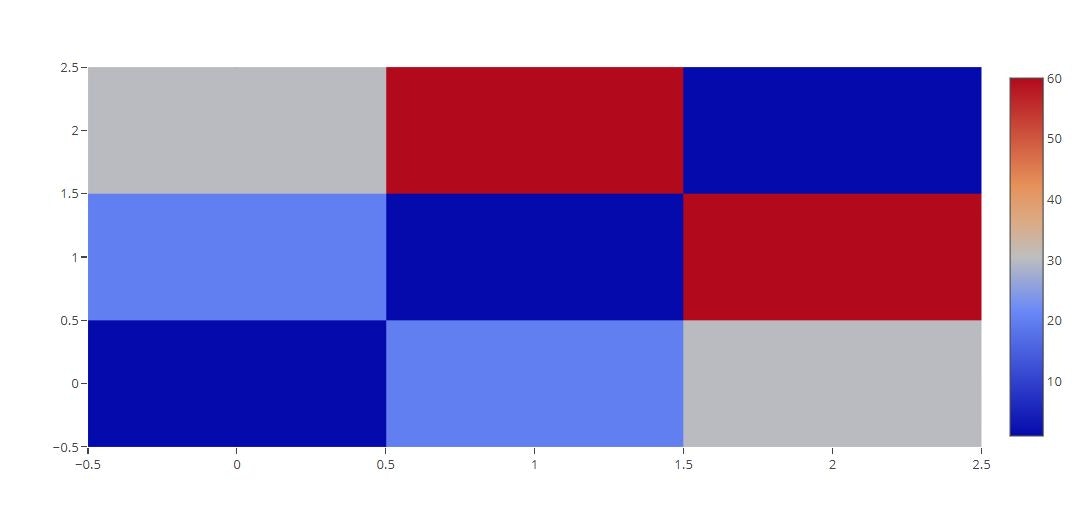
Heat Map
# Basic
trace = go.Heatmap(z=[[1, 20, 30],
[20, 1, 60],
[30, 60, 1]])
data=[trace]
py.offline.iplot(data)
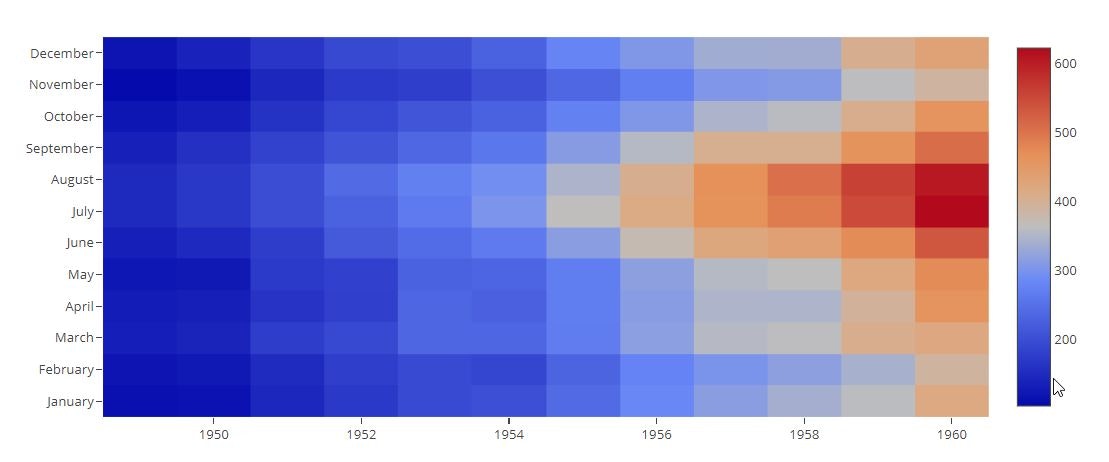
# Heatmap with Categorical Axis Labels
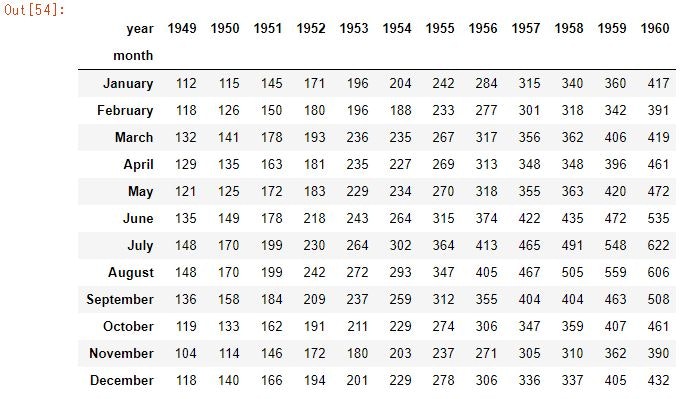
data = pd.pivot_table(data=flights_data, values='passengers', columns='year', index='month', aggfunc=np.mean)
data
trace = go.Heatmap(z=data.values,
x=data.columns,
y=data.index)
data=[trace]
py.offline.iplot(data)
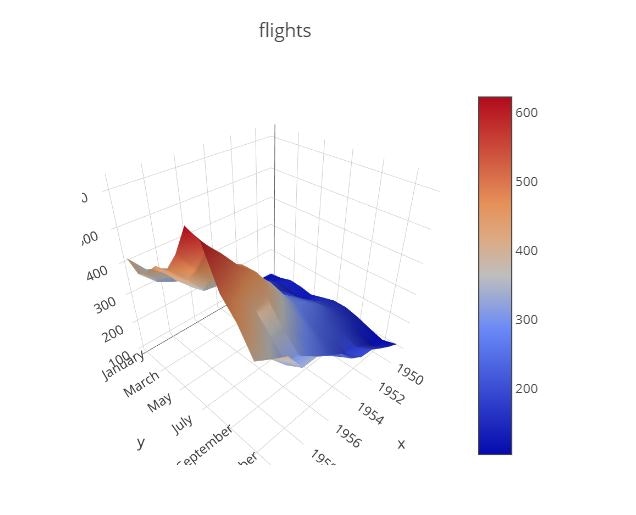
3D Surface
pivot_data = pd.pivot_table(data=flights_data, values='passengers', columns='year', index='month', aggfunc=np.mean)
data = [
go.Surface(
x=pivot_data.columns,
y=pivot_data.index,
z=pivot_data.values
)
]
layout = go.Layout(
title='flights',
autosize=False,
width=500,
height=500,
margin=dict(
l=65,
r=50,
b=65,
t=90
)
)
fig = go.Figure(data=data, layout=layout)
py.offline.iplot(fig)
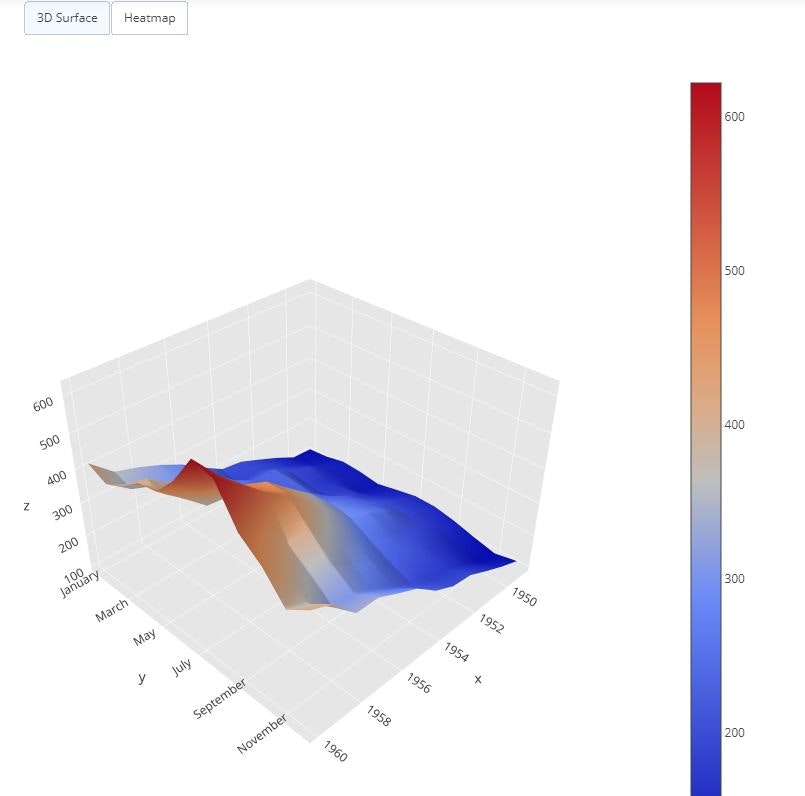
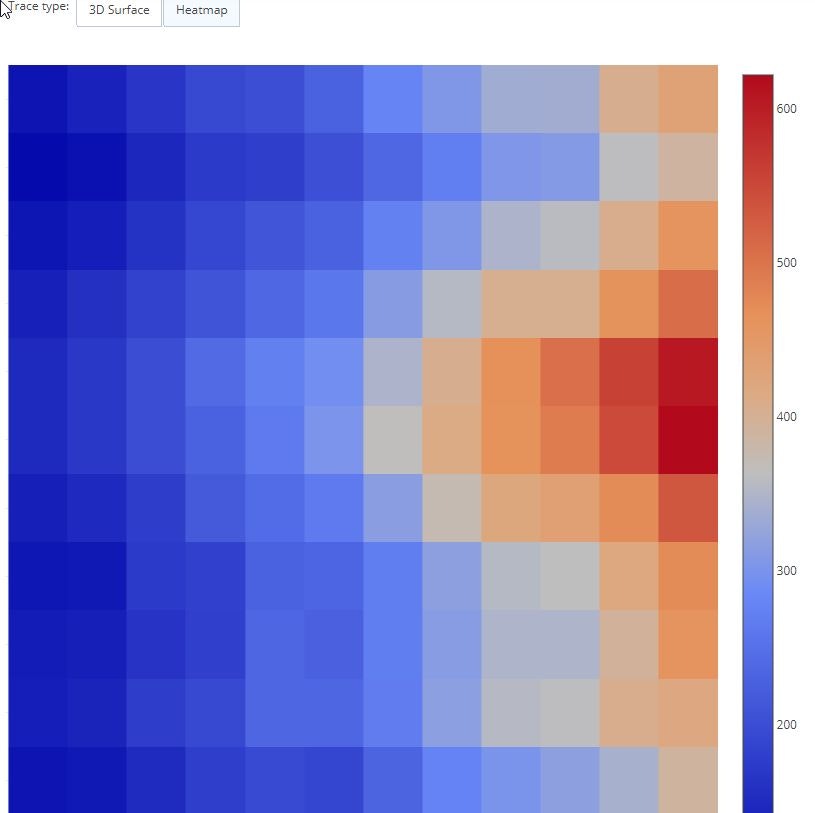
Restyle Button
pivot_data = pd.pivot_table(data=flights_data, values='passengers', columns='year', index='month', aggfunc=np.mean)
data = [
go.Surface(
x=pivot_data.columns,
y=pivot_data.index,
z=pivot_data.values
)
]
layout = go.Layout(
width=800,
height=900,
autosize=False,
margin=dict(t=0, b=0, l=0, r=0),
scene=dict(
xaxis=dict(
gridcolor='rgb(255, 255, 255)',
zerolinecolor='rgb(255, 255, 255)',
showbackground=True,
backgroundcolor='rgb(230, 230,230)'
),
yaxis=dict(
gridcolor='rgb(255, 255, 255)',
zerolinecolor='rgb(255, 255, 255)',
showbackground=True,
backgroundcolor='rgb(230, 230, 230)'
),
zaxis=dict(
gridcolor='rgb(255, 255, 255)',
zerolinecolor='rgb(255, 255, 255)',
showbackground=True,
backgroundcolor='rgb(230, 230,230)'
),
aspectratio = dict(x=1, y=1, z=0.7),
aspectmode = 'manual'
)
)
updatemenus=list([
dict(
buttons=list([
dict(
args=['type', 'surface'],
label='3D Surface',
method='restyle'
),
dict(
args=['type', 'heatmap'],
label='Heatmap',
method='restyle'
)
]),
direction = 'left',
pad = {'r': 10, 't': 10},
showactive = True,
type = 'buttons',
x = 0.1,
xanchor = 'left',
y = 1.1,
yanchor = 'top'
),
])
annotations = list([
dict(text='Trace type:', x=1949, y=1.085, yref='paper', align='left', showarrow=False)
])
layout['updatemenus'] = updatemenus
layout['annotations'] = annotations
fig = dict(data=data, layout=layout)
py.offline.iplot(fig)
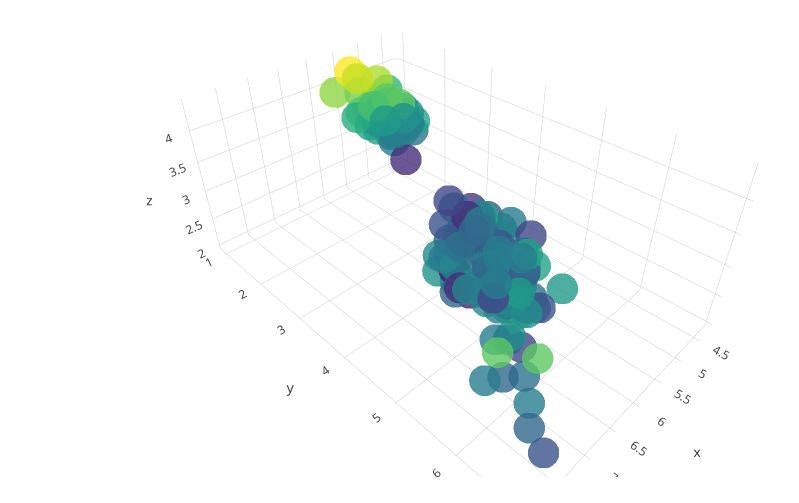
3D Scatter
trace1 = go.Scatter3d(
x=iris_data['sepal_length'],
y=iris_data['petal_length'],
z=iris_data['sepal_width'],
mode='markers',
marker=dict(
size=12,
color=iris_data['sepal_width'], # set color to an array/list of desired values
colorscale='Viridis', # choose a colorscale
opacity=0.8
)
)
data = [trace1]
layout = go.Layout(
margin=dict(
l=0,
r=0,
b=0,
t=0
)
)
fig = go.Figure(data=data, layout=layout)
py.offline.iplot(fig)
最後に
Plotlyは確かに綺麗なグラフが描けるんだけど、コーディングが結構煩雑になりやすい。
コーディングミスにも気を配る必要が出てくるから、特にこだわりが無いならSeaborn使った方が早いし無難かなと感じる。
会議とか見せ方を工夫したいときには、Plotly使うといいと思う。