研究会での論文読みにてTalevich, Eric, et al. "Bio. Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython." BMC bioinformatics 13.1 (2012): 1.を読んだので,そのメモをば.
Biopython とは
Bioinformatics用に開発されたPythonのライブラリ.Biopython Tutorial and Cookbook でその使い方が説明されている.
pip install biopython
Bio.Phylo
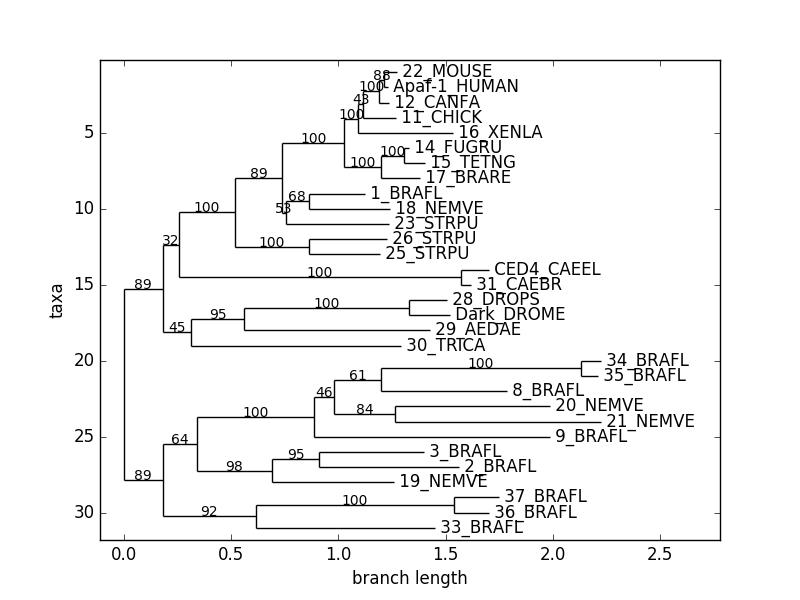
Bio.Phyloは系統樹の作成ができる.
論文中ではurllib2を使用しているがpython3ではurlopenはurllibに統合されている.
# coding: utf-8
from Bio import Phylo
from urllib.request import urlopen
handle = urlopen("http://www.phyloxml.org/examples/apaf.xml")
tree = Phylo.read(handle, "phyloxml")
handle.close()
Phylo.draw(tree)
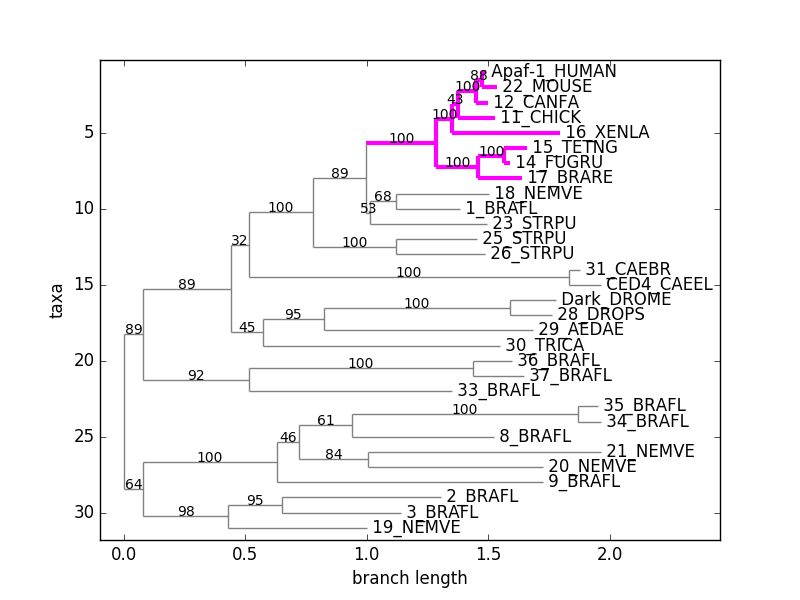
# coding: utf-8
from Bio import Phylo
from urllib.request import urlopen
handle = urlopen("http://www.phyloxml.org/examples/apaf.xml")
tree = Phylo.read(handle, "phyloxml")
handle.close()
tree.root_at_midpoint()
tree.ladderize(reverse=True)
vertebrata = tree.common_ancestor("Apaf-1_HUMAN", "15_TETNG")
vertebrata.color = "fuchsia"
vertebrata.width = 3
tree.root.color = "gray"
Phylo.draw(tree)
draw()の代わりにdraw_ascii()を用いると系統樹をアスキーアートとして出力してくれる.
_ 22_MOUSE
|
_| Apaf-1_HUMAN
| |
,| | 12_CANFA
||
_||___ 11_CHICK
| |
| |___________ 16_XENLA
_______|
| | , 14_FUGRU
| | __|
| |____| |__ 15_TETNG
_____| |
| | |____ 17_BRARE
| |
| | ______ 1_BRAFL
| | __|
______| || |_________ 18_NEMVE
| | |
| | |____________ 23_STRPU
| |
_| | _________ 26_STRPU
| | |_________|
| | |________ 25_STRPU
| |
| | ___ CED4_CAEEL
| |___________________________________|
____| |_ 31_CAEBR
| |
| | ___ 28_DROPS
| | _____________________|
| | ______| |____ Dark_DROME
| | | |
| |__| |_______________________ 29_AEDAE
| |
| |__________________________ 30_TRICA
|
| _ 34_BRAFL
| _________________________|
_| _____| |_ 35_BRAFL
| | |
| __| |_______________ 8_BRAFL
| | |
| | | ___________________ 20_NEMVE
| ______________| |_______|
| | | |__________________________ 21_NEMVE
| | |
| ___| |______________________________ 9_BRAFL
| | |
| | | _____________ 3_BRAFL
| | | _____|
| | |_________| |_________________ 2_BRAFL
|____| |
| |_______________ 19_NEMVE
|
| _____ 37_BRAFL
| ________________________|
|___________| |____ 36_BRAFL
|
|______________________ 33_BRAFL
参考文献
Talevich, Eric, et al. "Bio. Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython." BMC bioinformatics 13.1 (2012): 1.