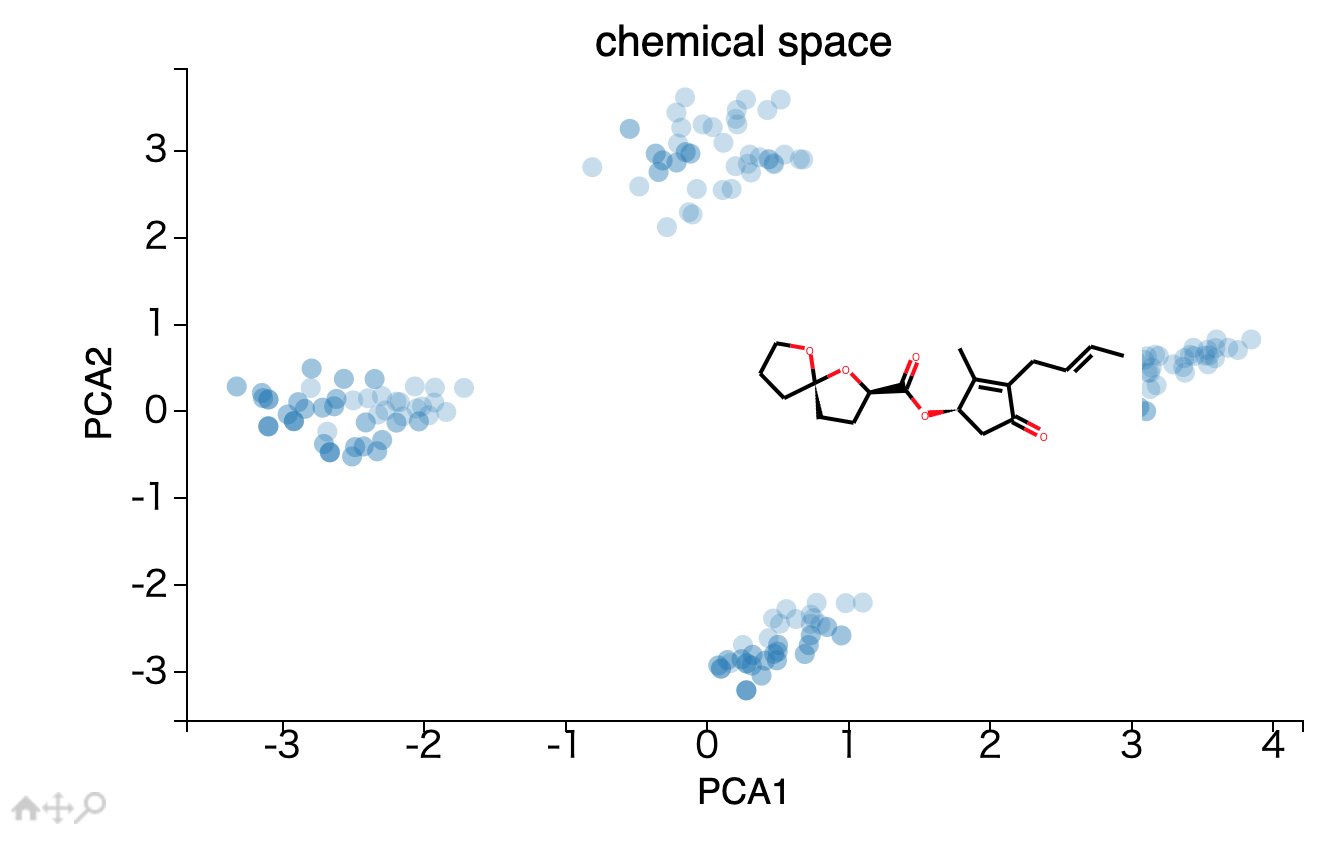
カーソルを当てると、構造式が表示される散布図の作り方
test.py
import pandas as pd
import numpy as np
from rdkit import Chem
from rdkit.Chem.Draw import rdDepictor
from rdkit.Chem.Draw import rdMolDraw2D
from rdkit.Chem import AllChem
from rdkit.Chem import DataStructs
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
from mpld3 import plugins
import mpld3
# mpld3.enable_notebook()
def main():
df = load_data('generated_mols.csv')
df['svg'] = [moltosvg(mol) for mol in df['mol']]
df = pca_for_df(df)
# print(df.head(20))
interactive_plot(df)
def load_data(file_name):
df = pd.read_csv(file_name,index_col=0,header=0)
df['mol'] = [Chem.MolFromSmiles(mol) for mol in df['SMILES']]
return df
def moltosvg(mol,molSize=(200,200),kekulize=True):
mc = Chem.Mol(mol.ToBinary())
if kekulize:
try:
Chem.Kekulize(mc)
except:
mc = Chem.Mol(mol.ToBinary())
if not mc.GetNumConformers():
rdDepictor.Compute2DCoords(mc)
drawer = rdMolDraw2D.MolDraw2DSVG(molSize[0],molSize[1])
drawer.DrawMolecule(mc)
drawer.FinishDrawing()
svg = drawer.GetDrawingText()
return svg.replace('svg:','')
def mol2fparr(mol):
arr = np.zeros((0,))
fp = AllChem.GetMorganFingerprintAsBitVect(mol,2)
DataStructs.ConvertToNumpyArray(fp, arr)
return arr
def pca_for_df(df):
pca = PCA(n_components=2)
X = np.asarray([mol2fparr(mol) for mol in df['mol']])
print(X.shape)
res = pca.fit_transform(X)
print(res.shape)
df['PCA1'] = res[:,0]
df['PCA2'] = res[:,1]
return df
def interactive_plot(df):
plt.rcParams["font.size"] = 18
fig, ax = plt.subplots(figsize=(10,6))
ax.set_xlabel('PCA1')
ax.set_ylabel('PCA2')
ax.set_title('chemical space')
points = ax.scatter(df['PCA1'], df['PCA2'],s=200,alpha=0.5,edgecolors='none')
tooltip = plugins.PointHTMLTooltip(points, df['svg'].values.tolist(),hoffset=10, voffset=10)
plugins.connect(fig, tooltip)
mpld3.save_html(fig, 'chemicalspace_for_generated_mols.html')
if __name__ == '__main__':
main()
SMILES式が入った下記のようなcsvファイルを用意しておく。
下記例では、すでにmolファイルの列があるが、SMILES式から作成するので不要。
generated_mols.csv
,mol,SMILES
0,<rdkit.Chem.rdchem.Mol object at 0x126093390>,C=CC=CCC1=C(C)[C@@H](OC2[C@H](OC)C2(C)C)CC1=O
1,<rdkit.Chem.rdchem.Mol object at 0x12609f3f0>,COC(=O)C(C)=C(C)C(=O)OC
2,<rdkit.Chem.rdchem.Mol object at 0x1263d34b0>,CO[C@H]1C(C(=O)O[C@H]2CC[C@@]3(CCCO3)O2)C1(C)C
3,<rdkit.Chem.rdchem.Mol object at 0x126467e10>,C=CC=C(C)C(=O)O[C@H]1C([C@H]2CC[C@@]3(CCCO3)O2)C1(C)C
4,<rdkit.Chem.rdchem.Mol object at 0x1263ccd50>,CC1(C)C(C(=O)O[C@H]2CC[C@@]3(CCCO3)O2)[C@@H]1[C@H]1CC[C@@]2(CCCO2)O1
5,<rdkit.Chem.rdchem.Mol object at 0x1263da870>,COC1[C@H](OC2[C@H]([C@H]3CC[C@@]4(CCCO4)O3)C2(C)C)C1(C)C
6,<rdkit.Chem.rdchem.Mol object at 0x125b126f0>,CC[C@H]1CC(=O)C(CC=CC2[C@H](CC)C2(C)C)=C1C
7,<rdkit.Chem.rdchem.Mol object at 0x1260b5150>,CC=C(C)C(=O)OC1[C@H](OC)C1(C)C
8,<rdkit.Chem.rdchem.Mol object at 0x126467570>,C=CC=C(C)C(=O)OC(=O)C(C)=CC=C