「StanとRでベイズ統計モデリング」 Chapter7より
交互作用
回帰分析において説明変数同士の掛け算の項を考慮すること
対数をとるか否か
モデル式7-1
$ \mu[n]= b_1+b_2Area[n] $
$ Y[n] \sim Normal(\mu[n],\sigma_Y) $
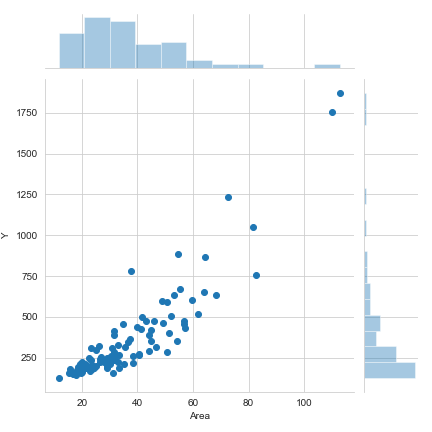
関東近郊の架空の賃貸物件データ
d = pd.read_csv("input/data-rental.txt")
sns.set_style("whitegrid")
sns.jointplot(x="Area",y="Y",data=d)
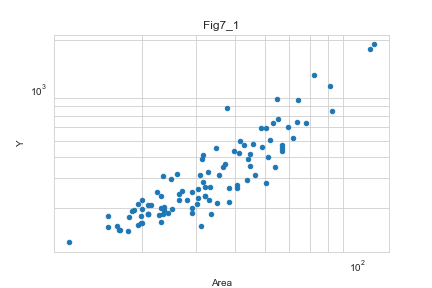
d.plot.scatter(x='Area',y='Y')
plt.yscale('log')
plt.xscale('log')
plt.grid(which="both")
plt.xlabel("Area")
plt.ylabel("Y")
sns.set_style('whitegrid')
plt.title('Fig7_1')
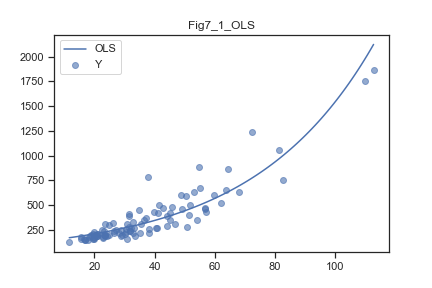
散布図:
横軸に説明変数、縦軸に応答変数
両対数軸でプロット
mdl = glm("Y~Area",data=d,family=sm.families.Poisson()).fit()
plt.scatter(d['Area'], d['Y'], alpha=0.6)
xx = np.linspace(min(d['Area']),max(d['Area']),100)
plt.plot(xx, np.exp(mdl.params[0]+mdl.params[1]*xx), 'b-', label='OLS')
plt.legend()
plt.title('Fig7_1_OLS')
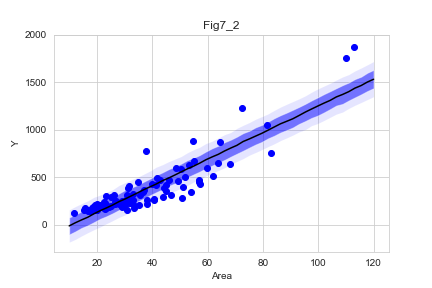
stanmodel = StanModel(file="model/model7-1.stan")
Area_new = np.linspace(10,120,50)
data_ = {'N':len(d),'Area':d['Area'],'Y':d['Y'],'N_new':50,'Area_new':Area_new}
fit1 = stanmodel.sampling(data=data_,n_jobs=-1,seed=1234)
ms1 = fit1.extract()
col = np.linspace(10,120,50)
df2 = pd.DataFrame(fit1['y_new'])
df2.columns = col
qua = [0.1, 0.25, 0.50, 0.75, 0.9]
d_est = pd.DataFrame()
for i in np.arange(len(df2.columns)):
for qu in qua:
d_est[qu] = df2.quantile(qu)
x = d_est.index
y1 = d_est[0.1].values
y2 = d_est[0.25].values
y3 = d_est[0.5].values
y4 = d_est[0.75].values
y5 = d_est[0.9].values
plt.fill_between(x,y1,y5,facecolor='blue',alpha=0.1)
plt.fill_between(x,y2,y4,facecolor='blue',alpha=0.5)
plt.plot(x,y3,'k-')
plt.scatter(d["Area"],d["Y"],c='b')
plt.xlabel("Area")
plt.ylabel("Y")
sns.set_style('whitegrid')
plt.title('Fig7_2')
d_ori = d
quantile = [10,50,90]
colname = ['p'+str(x) for x in quantile]
d_qua = pd.DataFrame(np.percentile(ms1["y_pred"],q=quantile,axis=0).T,columns=colname)
d_ = pd.concat([d_ori,d_qua],axis=1)
d0 = d_
palette = sns.color_palette()
fig = plt.figure(figsize=(10,8))
ax = fig.add_subplot(1,1,1)
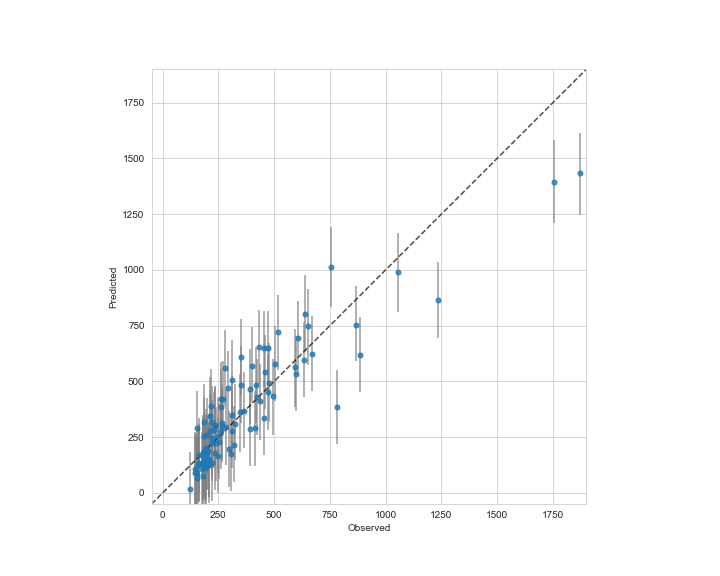
ax.plot([-50,1900],[-50,1900],'k--',alpha=0.7)
ax.errorbar(d0.Y,d0.p50,yerr=[d0.p50-d0.p10,d0.p90-d0.p50],
fmt='o',ecolor='gray',ms=5,mfc=palette[0],alpha=0.8,marker='o')
ax.set_aspect('equal')
ax.set_xlim(-50,1900)
ax.set_ylim(-50,1900)
ax.set_xlabel("Observed")
ax.set_ylabel("Predicted")
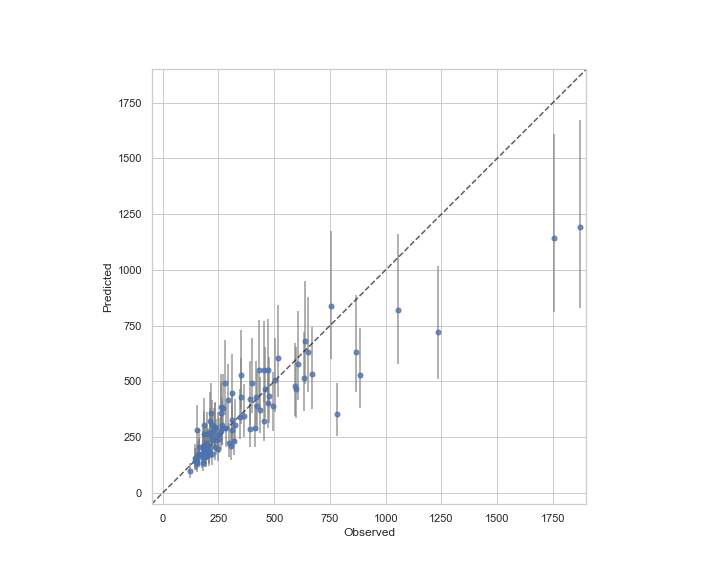
実測値と予測値のプロット
N_mcmc = len(ms1['lp__'])
d_noise = pd.DataFrame(-ms1['mu']+np.array(d_ori['Y']))
col = d_noise.columns
d_noise.columns = ["noise" + str(col[i]+1) for i in np.arange(len(col))]
data = d.copy()
data = data.sort_values(by="Area")
d_est = pd.concat([pd.DataFrame({"mcmc":np.arange(1,N_mcmc+1)}),d_noise],axis=1)
col = d_est.columns
est = []
fig = plt.figure(figsize=(10,8))
sns.set_style('whitegrid')
sns.set(font_scale=1)
for i in np.arange(1,len(d_est.columns)):
data = d_est[col[i]]
density = gaussian_kde(data)
xs = np.linspace(min(data),max(data),100)
plt.plot(xs,density(xs),'k-',lw=0.5)
xx = xs[np.argmax(density(xs))]
plt.vlines(xx,0,max(density(xs)),colors='k',linestyle="-.",alpha=0.5,linewidth=0.5)
est.append(xs[np.argmax(density(xs))])
plt.xlabel("Value")
plt.ylabel("density")
s_dens = gaussian_kde(ms1['s_Y'])
xs = np.linspace(min(ms1['s_Y']),max(ms1['s_Y']),100)
s_MPA = xs[np.argmax(s_dens(xs))]
print(s_MPA)
rv = norm(loc=0,scale=s_MPA)
bw = 25
bins = np.arange(min(est),max(est),bw)
fig = plt.figure(figsize=(10,8))
sns.set_style('whitegrid')
sns.set(font_scale=1)
res = plt.hist(est,bins=bins,color='lightgray',edgecolor='black')
x = np.linspace(rv.ppf(0.01),rv.ppf(0.99), 100)
plt.plot(x,rv.pdf(x)*len(d)*bw,'k--')
data = est
density = gaussian_kde(data)
print(max(data))
xs = np.linspace(min(data),max(data),100)
plt.plot(xs,density(xs)*len(d)*bw,lw=3)
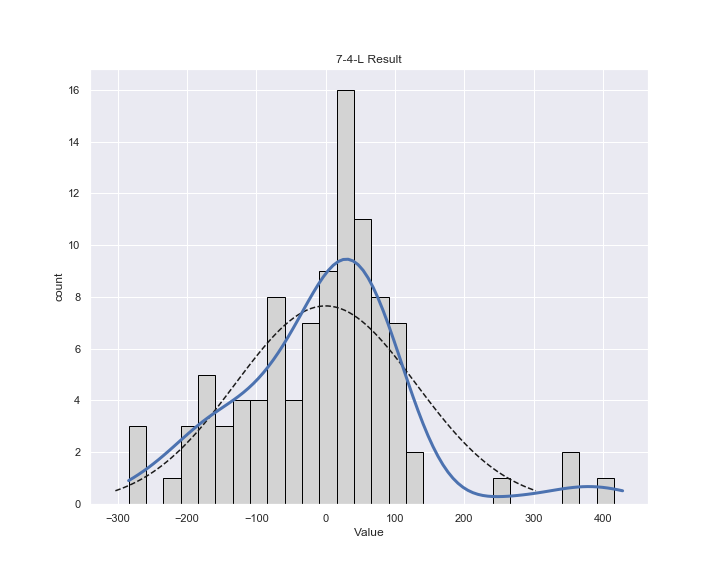
plt.title("7-4-L Result")
plt.xlabel("Value")
plt.ylabel("count")
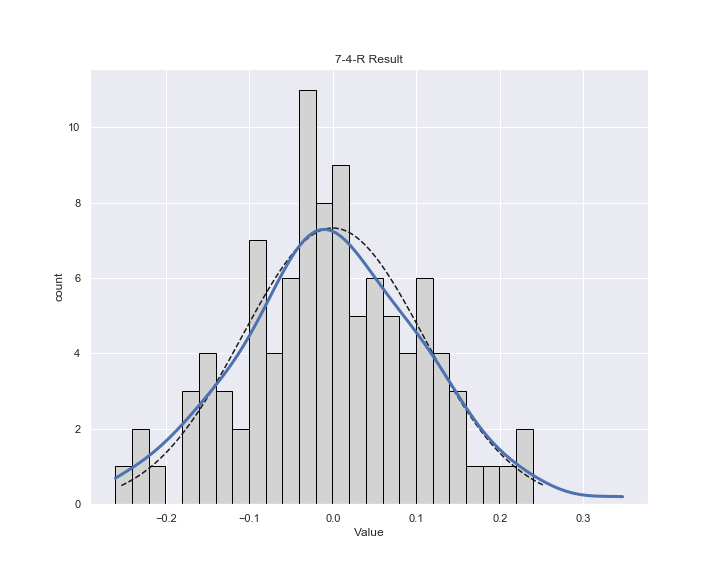
測定されたノイズの分布
広くて費用が高い物件(Areaがおよそ70m2以上の物件)に結果がひきずられてノイズの分布が歪んでいて、正規分布との乖離がある
モデル式7-2
YおよびAreaの対数をとって単回帰に使う
$ \mu[n]= b_1+b_2 log10(Area[n]) $
$ log10(Y[n]) \sim Normal(\mu[n],\sigma_Y) $
Area_new = np.linspace(10,120,50)
data_ = {'N':len(d),'Area':np.log10(d['Area']),'Y':np.log10(d['Y']),'N_new':50,'Area_new':np.log10(Area_new)}
fit2 = stanmodel.sampling(data=data_,n_jobs=-1,seed=1234)
ms2 = fit2.extract()
col = np.linspace(10,120,50)
df2 = pd.DataFrame(10**fit2['y_new'])
df2.columns = col
qua = [0.1, 0.25, 0.50, 0.75, 0.9]
d_est = pd.DataFrame()
for i in np.arange(len(df2.columns)):
for qu in qua:
d_est[qu] = df2.quantile(qu)
x = d_est.index
y1 = d_est[0.1].values
y2 = d_est[0.25].values
y3 = d_est[0.5].values
y4 = d_est[0.75].values
y5 = d_est[0.9].values
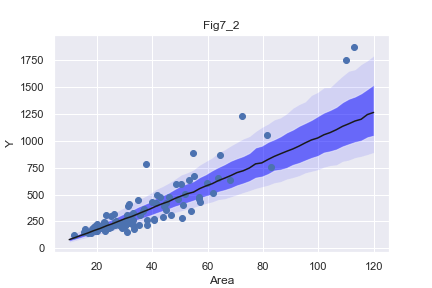
plt.fill_between(x,y1,y5,facecolor='blue',alpha=0.1)
plt.fill_between(x,y2,y4,facecolor='blue',alpha=0.5)
plt.plot(x,y3,'k-')
plt.scatter(d["Area"],d["Y"],c='b')
plt.xlabel("Area")
plt.ylabel("Y")
sns.set_style('whitegrid')
plt.title('Fig7_2')
d_ori = d
quantile = [10,50,90]
colname = ['p'+str(x) for x in quantile]
d_qua = pd.DataFrame(np.percentile(10**ms2["y_pred"],q=quantile,axis=0).T,columns=colname)
d_ = pd.concat([d_ori,d_qua],axis=1)
d0 = d_
palette = sns.color_palette()
fig = plt.figure(figsize=(10,8))
ax = fig.add_subplot(1,1,1)
ax.plot([-50,1900],[-50,1900],'k--',alpha=0.7)
ax.errorbar(d0.Y,d0.p50,yerr=[d0.p50-d0.p10,d0.p90-d0.p50],
fmt='o',ecolor='gray',ms=5,mfc=palette[0],alpha=0.8,marker='o')
ax.set_aspect('equal')
ax.set_xlim(-50,1900)
ax.set_ylim(-50,1900)
ax.set_xlabel("Observed")
ax.set_ylabel("Predicted")
Areaが1000m2より大きい物件のベイズ予測区間は広くなっている
N_mcmc = len(ms2['lp__'])
d_noise = pd.DataFrame(-ms2['mu']+np.array(np.log10(d_ori['Y'])))
col = d_noise.columns
d_noise.columns = ["noise" + str(col[i]+1) for i in np.arange(len(col))]
data = d.copy()
data = data.sort_values(by="Area")
d_est = pd.concat([pd.DataFrame({"mcmc":np.arange(1,N_mcmc+1)}),d_noise],axis=1)
col = d_est.columns
est = []
fig = plt.figure(figsize=(10,8))
sns.set_style('whitegrid')
sns.set(font_scale=1)
for i in np.arange(1,len(d_est.columns)):
data = d_est[col[i]]
density = gaussian_kde(data)
xs = np.linspace(min(data),max(data),100)
plt.plot(xs,density(xs),'k-',lw=0.5)
xx = xs[np.argmax(density(xs))]
plt.vlines(xx,0,max(density(xs)),colors='k',linestyle="-.",alpha=0.5,linewidth=0.5)
est.append(xs[np.argmax(density(xs))])
plt.xlabel("Value")
plt.ylabel("density")
s_dens = gaussian_kde(ms2['s_Y'])
xs = np.linspace(min(ms2['s_Y']),max(ms2['s_Y']),100)
s_MPA = xs[np.argmax(s_dens(xs))]
print(s_MPA)
rv = norm(loc=0,scale=s_MPA)
bw = 0.02
bins = np.arange(min(est),max(est),bw)
fig = plt.figure(figsize=(10,8))
sns.set_style('whitegrid')
sns.set(font_scale=1)
res = plt.hist(est,bins=bins,color='lightgray',edgecolor='black')
x = np.linspace(rv.ppf(0.01),rv.ppf(0.99), 100)
plt.plot(x,rv.pdf(x)*len(d)*bw,'k--')
data = est
density = gaussian_kde(data)
print(max(data))
xs = np.linspace(min(data),max(data),100)
plt.plot(xs,density(xs)*len(d)*bw,lw=3)
plt.title("7-4-R Result")
plt.xlabel("Value")
plt.ylabel("count")