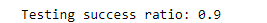Basis
Import packages
import matplotlib.pyplot as plt
import numpy as np
from qiskit import BasicAer
from qiskit.circuit.library import ZZFeatureMap
from qiskit.aqua import QuantumInstance, aqua_globals
from qiskit.aqua.algorithms import QSVM
from qiskit.aqua.utils import split_dataset_to_data_and_labels, map_label_to_class_name
seed = 10599
aqua_globals.random_seed = seed
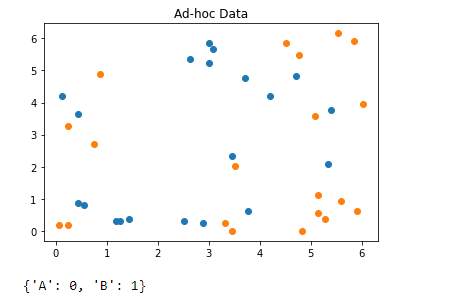
Dataset
(ad-hoc data)
from qiskit.ml.datasets import ad_hoc_data, sample_ad_hoc_data
feature_dim = 2
sample_total, training_input, test_input, class_labels = ad_hoc_data(
training_size=20,
test_size=10,
n=feature_dim,
gap=0.3,
plot_data=True
)
extra_test_data = sample_ad_hoc_data(sample_total, 10, n=feature_dim)
datapoints, class_to_label = split_dataset_to_data_and_labels(extra_test_data)
print(class_to_label)
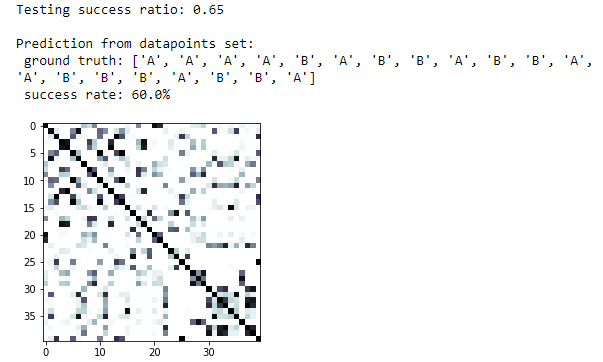
Success Ratio
feature_map = ZZFeatureMap(feature_dimension=feature_dim, reps=2, entanglement='linear')
qsvm = QSVM(feature_map, training_input, test_input, datapoints[0])
backend = BasicAer.get_backend('qasm_simulator')
quantum_instance = QuantumInstance(backend, shots=1024, seed_simulator=seed, seed_transpiler=seed)
result = qsvm.run(quantum_instance)
print(f'Testig success ratio: {result["testing_accuracy"]}')
print()
print('prediction from datapoints set:')
print(f' ground truth: {map_label_to_class_name(datapoints[1], qsvm.label_to_class)}')
print(f' prediction: {result["predicted_classes"]}')
predicted_labels = result["predicted_labels"]
print(f' success rate: {100*np.count_nonzero(predicted_labels == datapoints[1])/len(predicted_labels)}%')
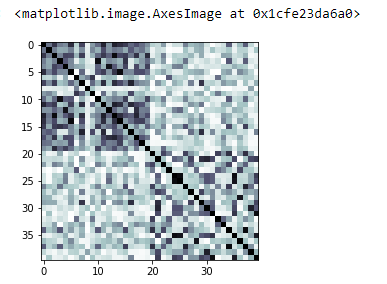
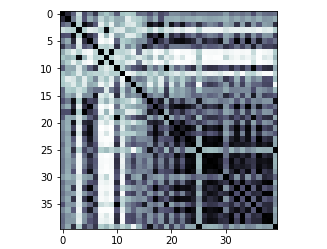
Kernel matrix that was built from the training sample of the dataset
kernel_matrix = result['kernel_matrix_training']
plt.imshow(
np.asmatrix(kernel_matrix),
interpolation='nearest',
origin="upper",
cmap='bone_r'
)
Classical SVM implementation that takes the same input data for classification
from qiskit.aqua.algorithms import SklearnSVM
result = SklearnSVM(training_input, test_input, datapoints[0]).run()
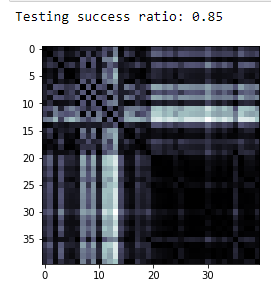
print(f'Testing success ratio: {result["testing_accuracy"]}')
print()
print('Prediction from datapoints set:')
print(f' ground truth: {result["predicted_classes"]}')
predicted_labels = result["predicted_labels"]
print(f' success rate: {100*np.count_nonzero(predicted_labels == datapoints[1])/len(predicted_labels)}%')
kernel_matrix = result['kernel_matrix_training']
plt.imshow(
np.asmatrix(kernel_matrix),
interpolation='nearest',
origin='upper',
cmap="bone_r"
);
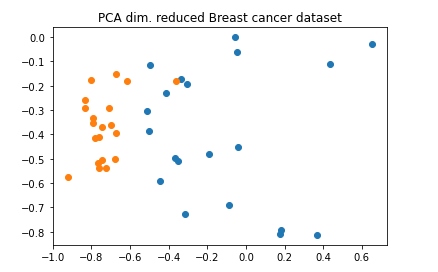
Practice by breast cancer data
from qiskit.ml.datasets import breast_cancer
feature_dim = 2
sample_total, training_input, test_input, class_labels = breast_cancer(
training_size=20,
test_size=10,
n=feature_dim,
plot_data=True
)
feature_map = ZZFeatureMap(feature_dimension=feature_dim, reps=2, entanglement='linear')
qsvm = QSVM(feature_map, training_input, test_input)
backend = BasicAer.get_backend('qasm_simulator')
quantum_instance = QuantumInstance(backend, shots=1024, seed_simulator=seed, seed_transpiler=seed)
result = qsvm.run(quantum_instance)
print(f'Testing success ratio: {result["testing_accuracy"]}')
kernel_matrix = result['kernel_matrix_training']
img = plt.imshow(np.asmatrix(kernel_matrix),interpolation='nearest',origin='upper',cmap='bone_r')
result = SklearnSVM(training_input, test_input).run()
print(f'Testing success ratio: {result["testing_accuracy"]}')
kernel_matrix = result['kernel_matrix_training']
plt.imshow(np.asmatrix(kernel_matrix), interpolation='nearest', origin='upper', cmap='bone_r');