library(ggplot2)
library(dplyr)
library(discretization)
binary_classification_plot_integer <- function(label, future, name){
# make df
df_marge <- data.frame(future=future,label=factor(label))
df_marge_mean <-
df_marge %>%
group_by(label) %>%
summarise(mean=mean(future)) %>%
as.data.frame
# plot
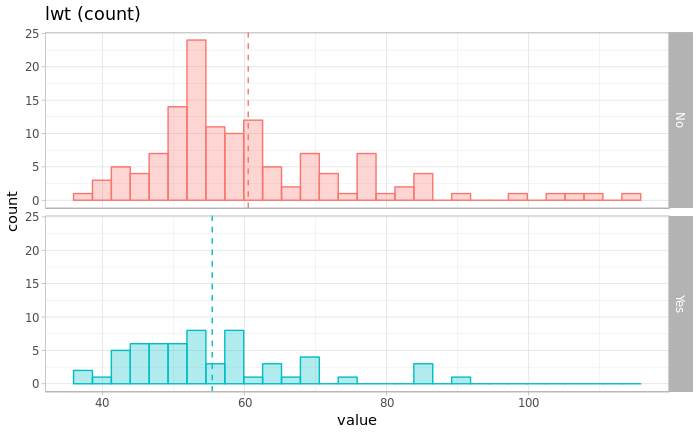
p <- ggplot(data=df_marge, aes(x=future, fill=label))
p <- p + geom_histogram(aes(colour=label),position="identity", alpha=0.3, bins=30)
p <- p + theme_light() + xlab("value") + ylab("count") + labs(title=paste0(name," (count)"))
p <- p + theme(legend.position="none")
p <- p + geom_vline(data=df_marge_mean, aes(xintercept=mean, color=label),linetype="dashed")
p <- p + facet_grid(label~.)
plot(p)
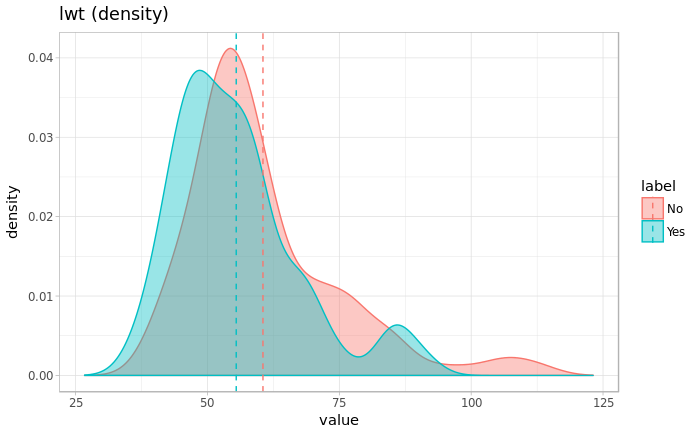
dens <- density(df_marge$future)
p <- ggplot(data=df_marge, aes(x=future, y=..density.., fill=label))
p <- p + geom_density(aes(colour=label), alpha =0.4)
p <- p + theme_light() + xlab("value") + ylab("density") + labs(title=paste0(name," (density)"))
p <- p + geom_vline(data=df_marge_mean, aes(xintercept=mean, color=label),linetype="dashed")
p <- p + xlim(range(dens$x))
plot(p)
# カイマージ
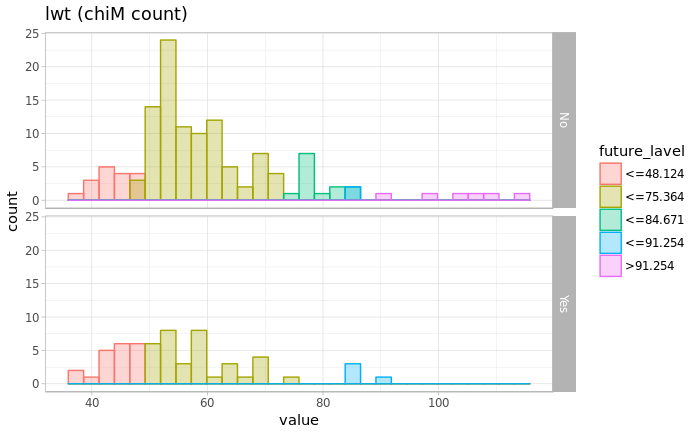
chiM <- chiM(df_marge, alpha=0.05)
df_chiM <- cbind(df_marge, future_class=chiM$Disc.data[,"future"])
# カイマージデータ
chiM_max <- max(df_chiM$future_class)
chiM_label <- c(paste0("<=",chiM$cutp[[1]]),paste0(">",chiM$cutp[[1]][chiM_max-1]))
chiM_master <- data.frame(future_class=c(1:chiM_max),future_lavel=chiM_label)
df_chiM_join <- inner_join(df_chiM, chiM_master ,by="future_class")
# カイマージの閾値を可視化
p <- ggplot(data=df_chiM_join, aes(x=future, fill=future_lavel))
p <- p + geom_histogram(aes(colour=future_lavel),position="identity", alpha=0.3, bins=30)
p <- p + theme_light() + xlab("value") + ylab("count") + labs(title=paste0(name," (chiM count)"))
p <- p + facet_grid(label~.)
plot(p)
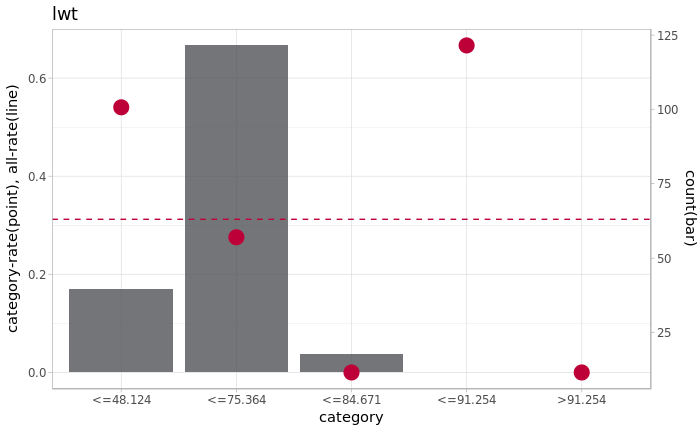
# カイマージのカテゴリの分布を可視化
binary_classification_plot_factor(df_chiM_join[,"label"], df_chiM_join[,"future_lavel"], name)
return(df_chiM_join)
}