「StanとRでベイズ統計モデリング」はワクワクする本なのですが、Rなのです。
随分前にRを使っていましたが、現在はpythonに乗り換えていますので、Rに戻るのは
気が乗りません。pythonで頑張りました。
以下のサイトを参考にしましたが、自分流に書いています。特に可視化のコードが分かりづらかったので、
pandasでseaborn用にデータ加工した後、seabornでポンと可視化しています。
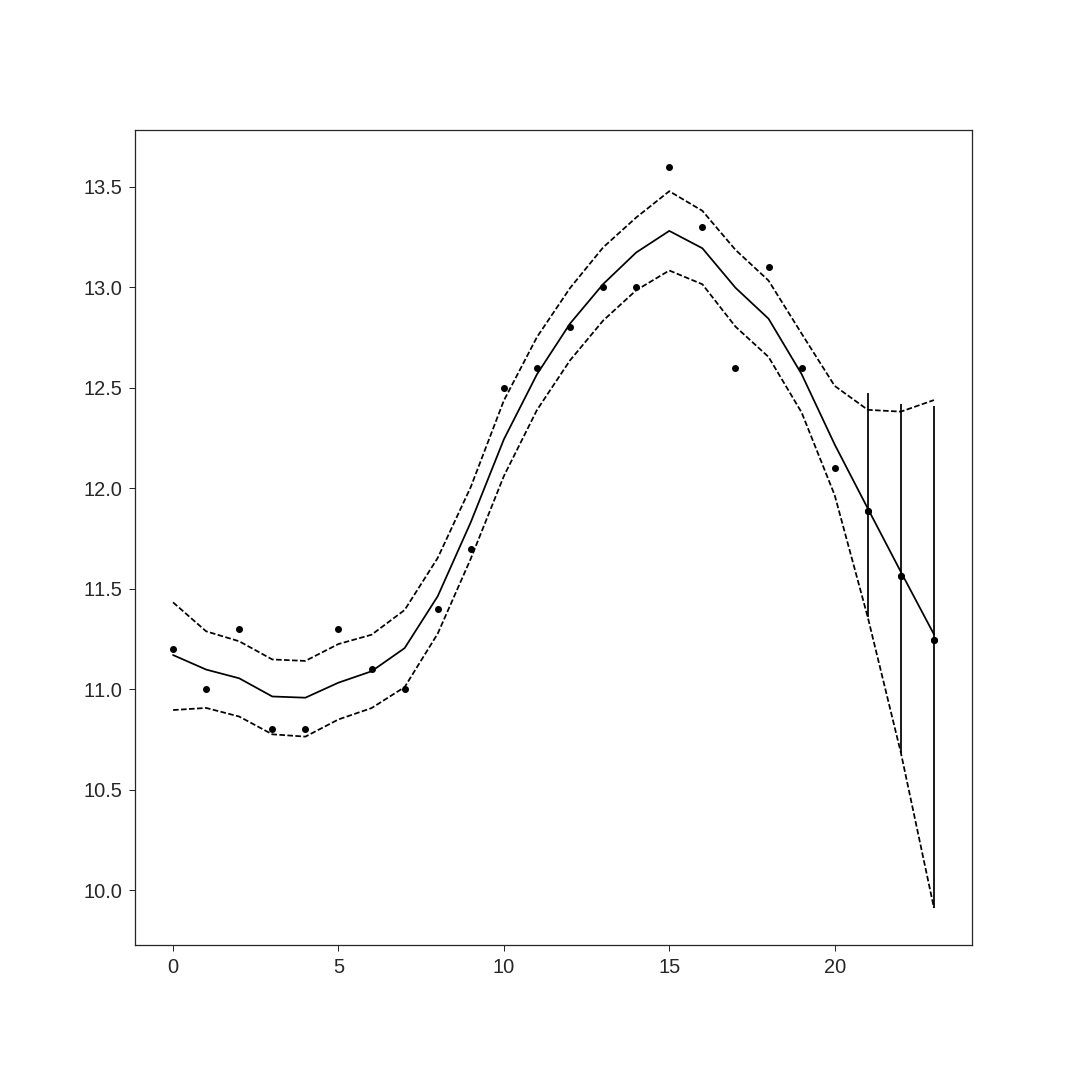
PyStan で「StanとRでベイズ統計モデリング」11.3節
Python(PyStan)で「StanとRでベイズ統計モデリング」の5.1節を実行する
状態空間モデルの可視化はRよりしょぼいです。Rのようにベイズ信頼区間を灰色にしたかったのですが、
できませんでした。
STAN良いですね! 夢が膨らみます!
seabornも良いです!
書籍のgithubはここです。
#!/usr/bin/env python
# -*- coding utf-8 -*-
# -------
# import
# -------
import os
import math
import pickle
import pystan
import functools
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
# ------------
# environment
# ------------
file_dir = os.getcwd()
# file_dir =os.path.abspath(os.path.dirname(__file__))
input_dir = os.path.join(file_dir, 'input')
model_dir = os.path.join(file_dir, 'model')
output_dir = os.path.join(file_dir, 'output')
data_file = os.path.join(input_dir, 'data-ss1.txt')
model_file = os.path.join(model_dir, 'model12-4.stan')
model_obj_file = os.path.join(output_dir, 'model.pkl')
fit_obj_file = os.path.join(output_dir, 'fit.pkl')
fit_file = os.path.join(output_dir, 'fit.txt')
# ---------
# compile
# ---------
model = pystan.StanModel(file=model_file)
with open(model_obj_file, 'wb') as f:
pickle.dump(model, f)
with open(model_obj_file, 'rb') as f:
model = pickle.load(f)
# ----------------
# data processing
# ----------------
data = pd.read_csv(data_file)
stan_data = {'T': len(data), 'T_pred': 3}
stan_data.update(data.to_dict('list'))
# ----------
# sampling
# ----------
fit = model.sampling(data=stan_data, n_jobs=1)
with open(fit_obj_file, 'wb') as f:
pickle.dump(fit, f)
with open(fit_file, 'w') as f:
f.write(str(fit))
with open(fit_obj_file, 'rb') as f:
fit = pickle.load(f)
# --------------------------
# prepare for visualization
# --------------------------
sample_wide = fit.extract(permuted=False, inc_warmup=False)
sample_wide = pd.DataFrame(
sample_wide.transpose(1,0,2).reshape(
sample_wide.shape[0] * sample_wide.shape[1],
sample_wide.shape[2]
),
columns=fit.sim['fnames_oi']
)
sample_long = sample_wide.unstack().reset_index()
sample_long = sample_long.drop('level_1', axis=1)
sample_long.columns = ['param', 'sample']
chain_cnt = fit.sim['chains']
sample_cnt = int(len(sample_wide) / fit.sim['chains'])
param_cnt = len(fit.sim['fnames_oi'])
sample_long['chain'] = (
np.ones((param_cnt, sample_cnt * chain_cnt), dtype='int64') *
np.repeat(range(0, chain_cnt), sample_cnt)
).ravel()
sample_long['seq'] =sample_long.groupby(['param', 'chain']).cumcount()
# ---------------
# sampling check
# ---------------
sns.set(font_scale=2)
sns.set_style("ticks")
sns.despine(offset=10, trim=True)
g = sns.FacetGrid(
sample_long,
row="param",
hue="chain",
size=5, aspect=3,
sharex=True, sharey=False
)
g = (g.map(plt.plot, "seq", "sample").add_legend())
plt.show()
sns.set(font_scale=2)
sns.set_style("ticks")
sns.despine(offset=10, trim=True)
g = sns.FacetGrid(
sample_long,
col="param",
col_wrap=9,
hue="chain",
size=3, aspect=1,
sharex=False, sharey=False
)
g = (g.map(sns.kdeplot, "sample").add_legend())
plt.show()
#sample_wide = sample_long[['seq', 'param', 'sample']].pivot_table(
# index='seq', columns='param', values='sample'
#)
g = sns.PairGrid(sample_wide[[
'mu_all[0]', 'mu_all[11]', 'mu_all[23]',
'y_pred[0]', 'y_pred[1]', 'y_pred[2]',
's_mu', 's_Y'
]])
g.map_upper(plt.scatter)
g.map_lower(sns.kdeplot, cmap="Blues_d")
g.map_diag(sns.distplot, hist=False, fit=stats.norm);
plt.show()
# --------------
# visualization
# --------------
state = pd.DataFrame(
np.percentile(fit.extract()['mu_all'], q=[10, 50, 90], axis=0).T,
columns=['state_{0}'.format(c) for c in ['lower', 'middle', 'upper']]
)
pred = pd.DataFrame(
np.percentile(fit.extract()['y_pred'], q=[10, 50, 90], axis=0).T,
columns=['pred_{0}'.format(c) for c in ['lower', 'middle', 'upper']]
)
data['T'] = range(0, len(data))
state['T'] = range(0, len(state))
pred['T'] = range(21, 24)
result = functools.reduce(
lambda left, right: pd.merge(left, right, on='T', how='left'), (
state,
pred,
data[['T', 'Y']]
)
)
sns.set(font_scale=2)
sns.set_style("ticks")
sns.despine(offset=10, trim=True)
plt.figure(figsize=(15, 15))
plt.plot(result['T'], result['Y'], 'o', color='black')
plt.plot(result['T'], result['state_middle'], '-', color='black')
plt.plot(result['T'], result['state_upper'], '--', color='black')
plt.plot(result['T'], result['state_lower'], '--', color='black')
plt.plot(result['T'], result['pred_middle'], 'o', color='black')
plt.errorbar(
result['T'], result['pred_middle'],
yerr=[
result['pred_middle'] - result['state_lower'],
result['pred_upper'] - result['state_middle']
],
color='black', ecolor='black', fmt='o'
)
plt.show()