RDkitのインストール
2015/08/23現在
python3用rdkitはcondaを使ってbuild可能
参考URL
conda install conda-build
git clone https://github.com/rdkit/conda-rdkit.git
cd conda-rdkit/
git checkout development
CONDA_PY=34 conda build boost
CONDA_PY=34 conda build rdkit
conda install rdkit --use-local
# 仮想環境に入れることも可能
# conda create --use-local -n py3_rdkit python=3.4 pip rdkit
rdkitデモ
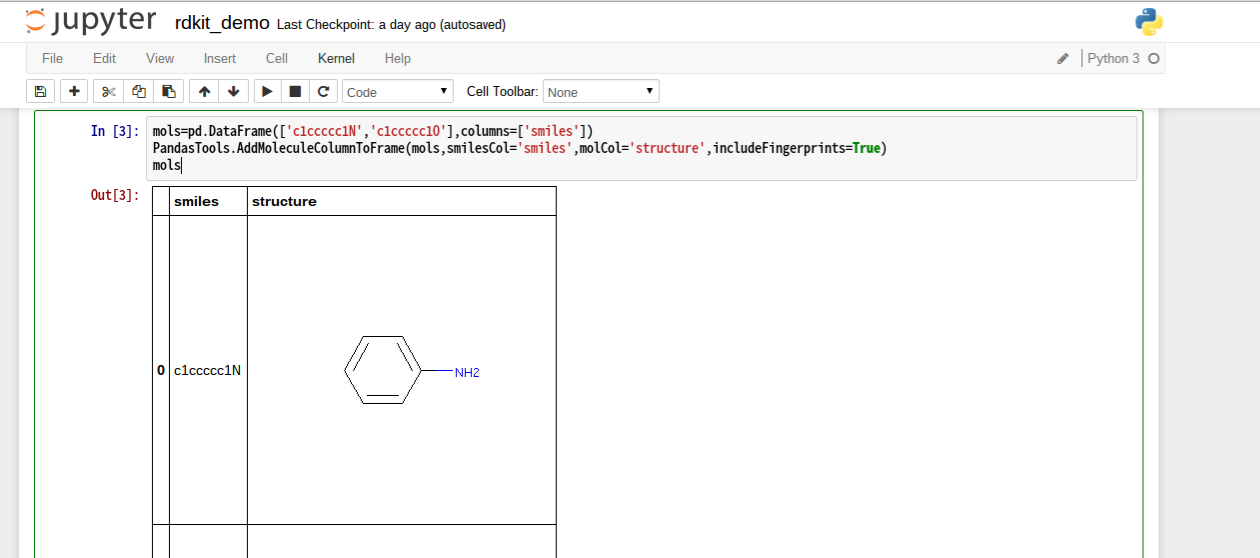
smilesからpd.DataFrameに読み込む
# coding: utf-8
# In[1]:
import pandas as pd
from numpy import vectorize as vec
# In[2]:
from rdkit import Chem
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import Descriptors,PandasTools
# In[3]:
mols=pd.DataFrame(['c1ccccc1N','c1ccccc1O'],columns=['smiles'])
PandasTools.AddMoleculeColumnToFrame(mols,smilesCol='smiles',molCol='structure',includeFingerprints=True)
# sdfから読むときは
# PandasTools.LoadSDF(sdfFile,smilesName='SMILES',molColName='Molecule',includeFingerprints=True)
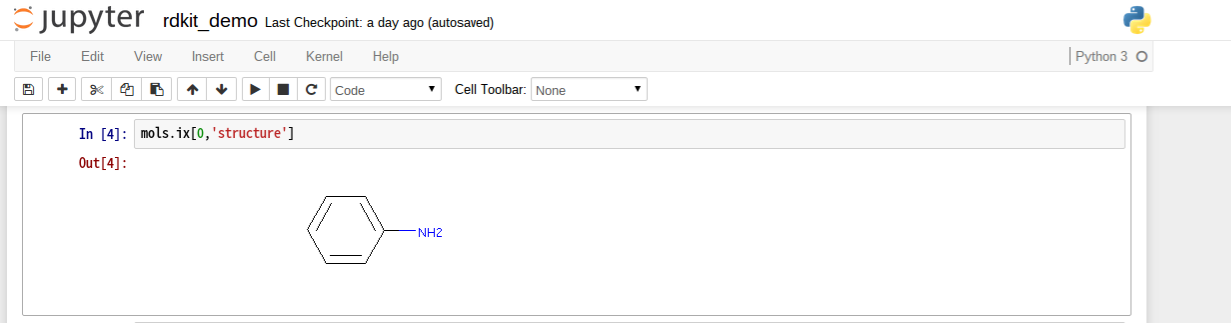
構造インライン表示
# In[4]:
mols.ix[0,'structure']
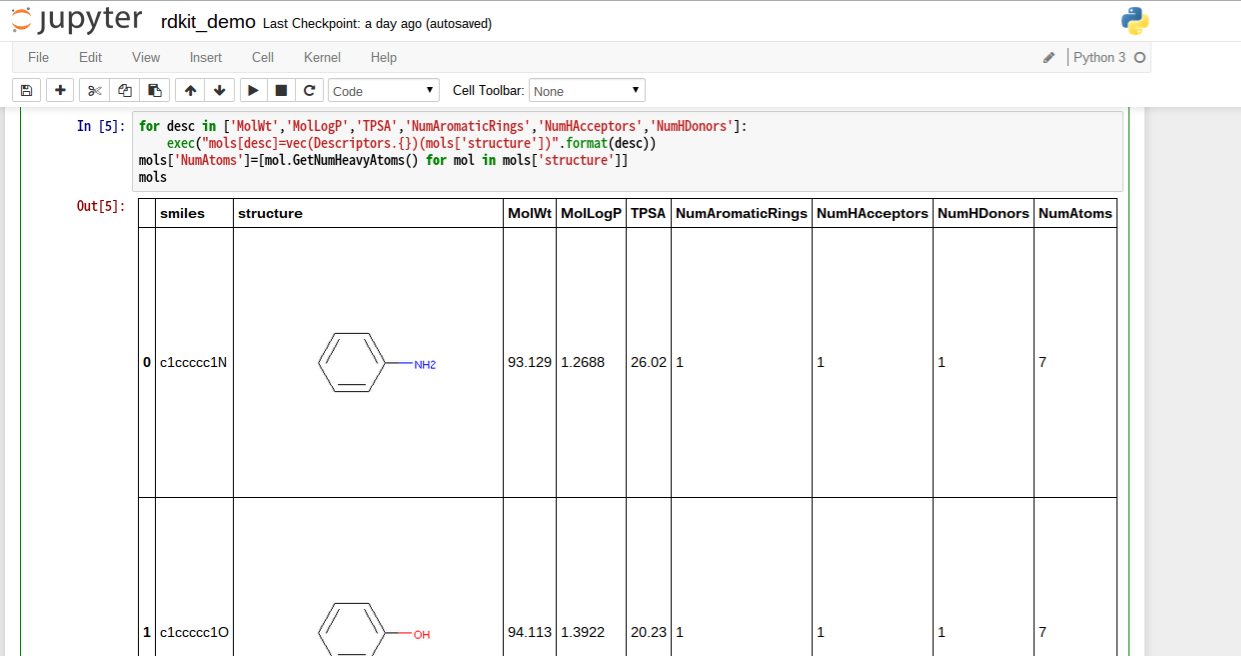
記述子計算
# In[5]:
for desc in ['MolWt','MolLogP','TPSA','NumAromaticRings','NumHAcceptors','NumHDonors']:
exec("mols[desc]=vec(Descriptors.{})(mols['structure'])".format(desc))
mols['NumAtoms']=[mol.GetNumHeavyAtoms() for mol in mols['structure']] #重原子数はmoleculeのmethodに定義されている
mols
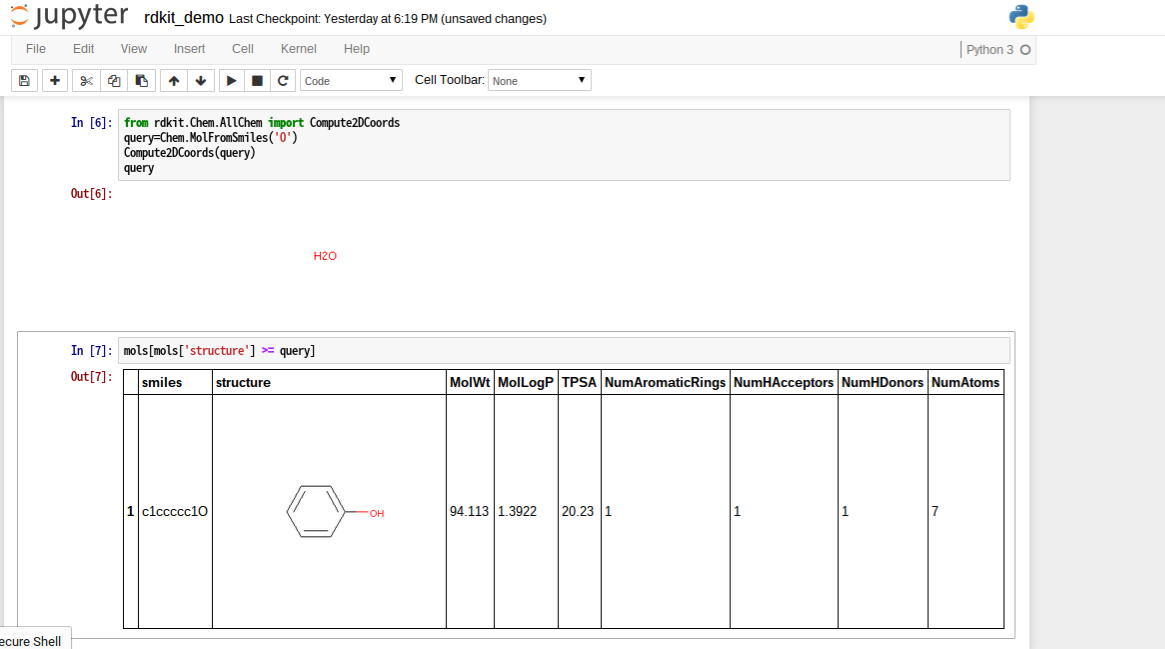
部分構造検索
# In[6]:
from rdkit.Chem.AllChem import Compute2DCoords
query=Chem.MolFromSmiles('O')
Compute2DCoords(query)
query
# In[7]
mols[mols['structure'] >= query]