動作環境
GeForce GTX 1070 (8GB)
ASRock Z170M Pro4S [Intel Z170chipset]
Ubuntu 14.04 LTS desktop amd64
TensorFlow v0.11
cuDNN v5.1 for Linux
CUDA v8.0
Python 2.7.6
IPython 5.1.0 -- An enhanced Interactive Python.
gcc (Ubuntu 4.8.4-2ubuntu1~14.04.3) 4.8.4
数値光散乱シミュレーションで出力されるチェックポイントファイルの読込みをCで実装した。
http://qiita.com/7of9/items/0fbea38b48cdfe4588dd
これをmatplotlibで読むためにPython実装に変更しようとしている。
http://qiita.com/7of9/items/4aa9f546a4ff1bf2e1aa
の続き。
v0.1
Jupyterに環境を移した。
あとで探せなくなるのでdisp_checkpoint.ipynbという記載を以下につけておく。
disp_checkpoint.ipynb
'''
disp_checkpoint.ipynb
'''
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import array
import sys
'''
v0.1 Dec. 23, 2016
- show plot of real part of xvec[]
+ add save_to_file()
branched from [read_chpoint.py] to Jupyter environment
v0.6 Dec. 23, 2016
- try to print fractional part of xvec[] in 4 digits
+ xvec[idx] = np.array([drel, dimg]), also for rvec[], pvec[], vectors[][]
+ add [np.set_printoptions(precision=4)]
v0.5 Dec. 23, 2016
- read scalars[], xvec[], rvec[], pvec[], vectors[][]
- refactor according to PEP8 (except for E501 line too long for long comment)
- wrap_fromfile() takes [num] arg
v0.4 Dec. 21, 2016
- read from "auxiliary" file [auxfp]
+ add read_auxiliary_info()
v0.3 Dec. 19, 2016
- add wrap_fromfile()
- read [inprodR],[prev_err],[resid_scale]
v0.2 Dec. 19, 2016
- fix bug > read [local_nRows] for "size_t" type
v0.1 Dec. 18, 2016
- read_chpoint_file() > read [ind_m],[local_nRows],[niter],[counter]
'''
np.set_printoptions(precision=6)
def wrap_fromfile(rfp, typecode, num):
res = array.array(typecode)
res.fromfile(rfp, num)
return res
def read_auxiliary_info(auxFilename):
with open(auxFilename, "rb") as auxfp:
sc_N = wrap_fromfile(auxfp, 'i', num=1) # the number of scalars' sizes
sc_sizes = array.array('i') #
sc_sizes.fromfile(auxfp, *sc_N) # '*':unpack list
vec_N = wrap_fromfile(auxfp, 'i', num=1) # the number of vercotrs' sizes
vec_sizes = array.array('i') #
vec_sizes.fromfile(auxfp, *vec_N) # '*':unpack list
return sc_N, sc_sizes, vec_N, vec_sizes
# TODO: 0m > error handling when file is NULL
def save_to_file(xvec, local_nRows):
data_1d = []
print(local_nRows[0])
#for idx in range(local_nRows[0]):
for idx in range(167*167): # original size 27889=local_nRows[0]
data_1d.append(*xvec[idx][0]) # real part of xvec[]
data_2d = np.reshape(data_1d, (167,167))
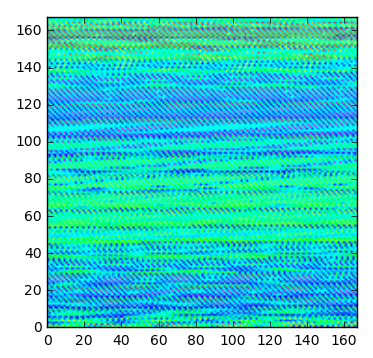
plt.imshow(data_2d, extent=(0,167,0,167),cmap=cm.gist_rainbow)
plt.show()
def read_chpoint_file(chpFilename, auxFilename):
print(chpFilename)
print(auxFilename)
# TODO: 0m > error handling when file is NULL
with open(chpFilename, "rb") as chpfp:
ind_m = wrap_fromfile(chpfp, 'i', num=1) # index of iterative method
print('ind_m:', ind_m)
# number of local rows of decomposition (only real dipoles)
# where L: unsigned long = size_t
local_nRows = wrap_fromfile(chpfp, 'L', num=1)
print('local_nRows:', local_nRows)
niter = wrap_fromfile(chpfp, 'i', num=1) # iteration count
print('niter:', niter)
# number of successive iterations without residual decrease
counter = wrap_fromfile(chpfp, 'i', num=1)
print('counter:', counter)
# used as |r_0|^2 and best squared norm of residual up to some iteration
inprodR = wrap_fromfile(chpfp, 'd', num=1)
print('inprodR:', inprodR)
# previous relative error; used in ProgressReport, initialized in IterativeSolver
prev_err = wrap_fromfile(chpfp, 'd', num=1)
print('prev_err:', prev_err)
# scale to get square of relative error
resid_scale = wrap_fromfile(chpfp, 'd', num=1)
print('resid_scale:', resid_scale) # scale to get square of relative error
# sc_N: the number of scalars' sizes
# sc_sizes:
# vec_N: the number of vercotrs' sizes
# vec_sizes:
sc_N, sc_sizes, vec_N, vec_sizes = read_auxiliary_info(auxFilename)
print('sc_N:', sc_N)
print('sc_sizes', sc_sizes)
print('vec_N:', vec_N)
print('vec_sizes:', vec_sizes)
# scalars[]
scalars = []
for idx in range(sc_N[0]):
if sc_sizes[idx] is 8: # double
scl = wrap_fromfile(chpfp, 'd', num=1)
scalars.append(scl)
elif sc_sizes[idx] is 16: # double complex
scl = wrap_fromfile(chpfp, 'd', num=2)
scalars.append(scl)
for idx in range(sc_N[0]):
print('scalars:', scalars[idx])
# xvec[] : total electric field on the dipoles
# TODO: 0m > replace 'local_nRows[0]' with function?
# TODO: 0m > replace 'vec_N[0]' with function?
# TODO: 0m > define function for read [drel,dimg]??
xvec = [[] for idx in range(local_nRows[0])]
for idx in range(local_nRows[0]):
drel = wrap_fromfile(chpfp, 'd', 1)
dimg = wrap_fromfile(chpfp, 'd', 1)
xvec[idx] = np.array([drel, dimg])
# rvec[] : current residual
rvec = [[] for idx in range(local_nRows[0])]
for idx in range(local_nRows[0]):
drel = wrap_fromfile(chpfp, 'd', 1)
dimg = wrap_fromfile(chpfp, 'd', 1)
rvec[idx] = np.array([drel, dimg] )
# pvec[] : polarization of dipoles, also an auxiliary vector in iterative solvers
pvec = [[] for idx in range(local_nRows[0])]
for idx in range(local_nRows[0]):
drel = wrap_fromfile(chpfp, 'd', 1)
dimg = wrap_fromfile(chpfp, 'd', 1)
pvec[idx] = np.array([drel, dimg])
# vectors[] : ???
MAXNUM_VECTORS = 20
vectors = [[] for idx in range(MAXNUM_VECTORS)]
for idx_vecN in range(vec_N[0]):
alist = []
for idx_LnRows in range(local_nRows[0]):
if vec_sizes[idx_vecN] is 16:
drel = wrap_fromfile(chpfp, 'd', 1)
dimg = wrap_fromfile(chpfp, 'd', 1)
alist.append(np.array([drel, dimg]))
vectors[idx_vecN] = alist
#
for idx in range(3):
print('xvec:',np.array_repr(xvec[idx]).replace('\n',''))
print('rvec:',np.array_repr(rvec[idx]).replace('\n',''))
print('pvec:',np.array_repr(pvec[idx]).replace('\n',''))
print('vectors:',np.array_repr(vectors[idx][0]).replace('\n',''))
print('vectors:',np.array_repr(vectors[idx][1]).replace('\n',''))
print('vectors:',np.array_repr(vectors[idx][2]).replace('\n',''))
#
return xvec, local_nRows
argvs = sys.argv
argc = len(argvs)
print argvs
if (argc < 3):
print("ERROR: chpoint file is not specified\r\n")
print(" [cmd] [chpoint file] [auxiliary file]\r\n")
sys.exit()
# read_chpoint_file(chpFilename=argvs[1], auxFilename=argvs[2])
xvec, local_nRows = read_chpoint_file("LN-CHP", "LN-AUX")
save_to_file(xvec, local_nRows)
結果
['/home/yasokada/tensorflow-GPU/lib/python2.7/site-packages/ipykernel/__main__.py', '-f', '/run/user/1000/jupyter/kernel-bbf67d4b-dda7-4d2b-b1ae-cd5205ce660d.json']
LN-CHP
LN-AUX
('ind_m:', array('i', [5]))
('local_nRows:', array('L', [27984L]))
('niter:', array('i', [209]))
('counter:', array('i', [139]))
('inprodR:', array('d', [4.317011265934948]))
('prev_err:', array('d', [0.3198525404317458]))
('resid_scale:', array('d', [0.020384616751203153]))
('sc_N:', array('i', [8]))
('sc_sizes', array('i', [8, 8, 8, 8, 16, 16, 16, 16]))
('vec_N:', array('i', [3]))
('vec_sizes:', array('i', [16, 16, 16]))
('scalars:', array('d', [26.040533833425627]))
('scalars:', array('d', [21.695179646708983]))
('scalars:', array('d', [0.01879089326833417]))
('scalars:', array('d', [0.01622964090344006]))
('scalars:', array('d', [2.393088070369695, 5.705068471314085]))
('scalars:', array('d', [0.2463390807093872, 0.2808495718014472]))
('scalars:', array('d', [0.8838701376516526, -0.4673547711298418]))
('scalars:', array('d', [0.34430517188699605, -0.9387175013645006]))
('xvec:', 'array([[ 0.000916], [-0.001571]])')
('rvec:', 'array([[-0.0021 ], [ 0.002827]])')
('pvec:', 'array([[-0.001287], [-0.008525]])')
('vectors:', 'array([[-0.203491], [ 0.033598]])')
('vectors:', 'array([[ 0.513358], [-0.235685]])')
('vectors:', 'array([[ 0.548095], [ 0.061967]])')
('xvec:', 'array([[ 0.032771], [ 0.090957]])')
('rvec:', 'array([[ 0.004384], [-0.023016]])')
('pvec:', 'array([[-0.00586 ], [ 0.025872]])')
('vectors:', 'array([[ 0.100941], [-0.212583]])')
('vectors:', 'array([[-0.220141], [ 0.644602]])')
('vectors:', 'array([[-0.386403], [ 0.429201]])')
('xvec:', 'array([[-0.004613], [-0.000545]])')
('rvec:', 'array([[ 0.005385], [-0.006732]])')
('pvec:', 'array([[-0.000544], [ 0.022825]])')
('vectors:', 'array([[-0.000433], [ 0.008886]])')
('vectors:', 'array([[ 0.007084], [-0.024328]])')
('vectors:', 'array([[ 0.006453], [-0.022867]])')
27984